Innovations in scientific publishing
There are awesome things happening and changing around how science is done and communicated, so I thought I'd put them in a list thingy. Let's go!
Code Ocean 🌊
When I first saw it used, linked from a Nature paper, I thought it was a demo and not a real, living thing. It took me a good while to realise that it is, in fact, an actual product, and an awesome one at that. I mean just take a look at this https://codeocean.com/capsule/6314882/tree/v1, for example.
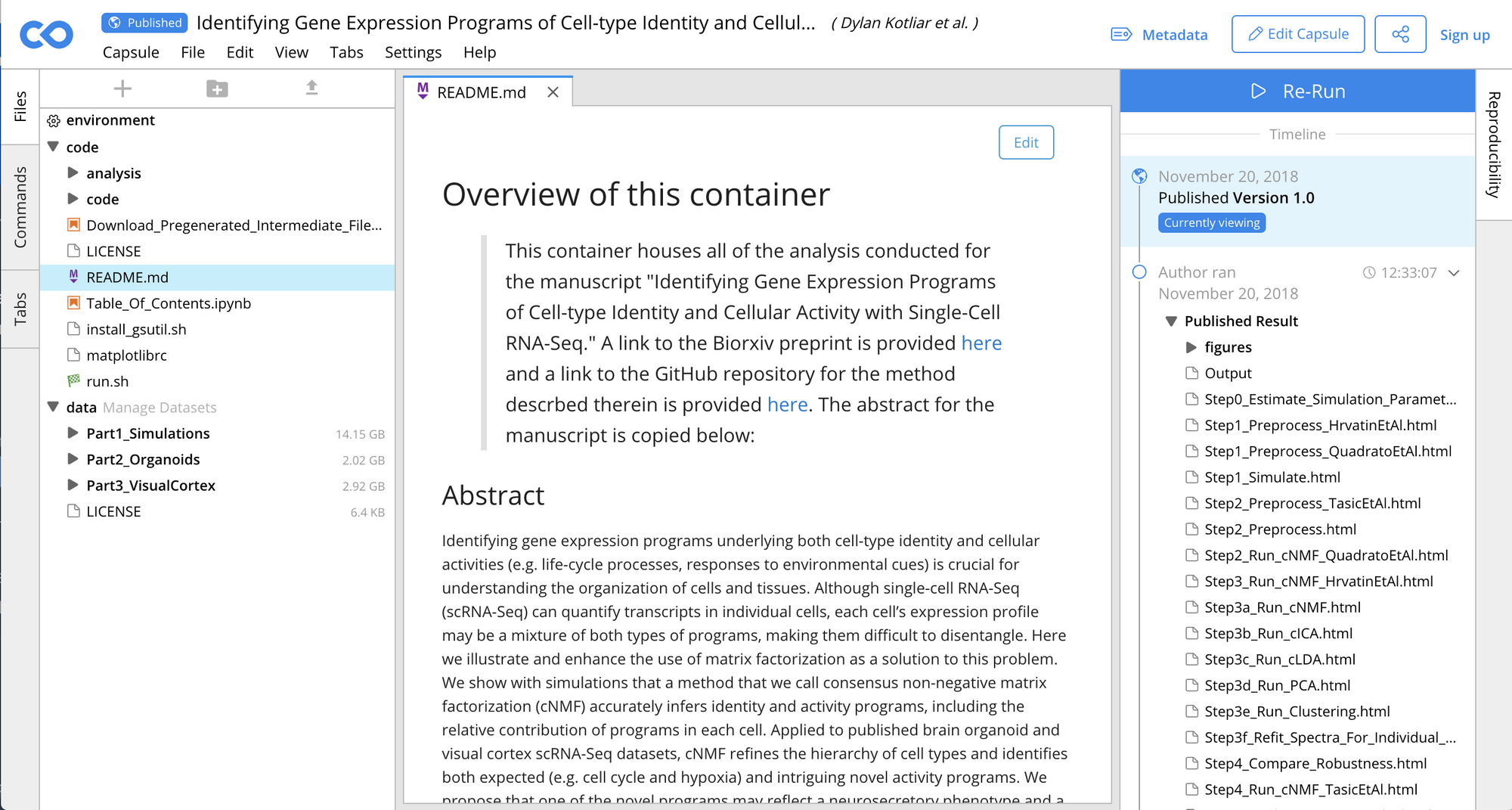
Just like that, there's the readme, there's the code that analyses and visualises the data, the data itself, the results, and clearly marked versions of it all. There's also a big blue "Computer, run the world, again" button if you want to test the reproducibility of this analysis yourself. We've been talking about this for years and here it is. Simply put, this is amazing. Integrated with R, Jupyter, indexed by search engines, archived by CLOCKSS, citable with a DOI, aah, I could go on and on!
Head over to https://codeocean.com/ to find out more about this giant step forward for reproducible open science. I only wish it were, well... open (source).
eLife's new submission system
One of my favourite open access journals, eLife, is developing a new submission system. Full disclosure, I'm a bit biased here, since they're also using technology that the PubSweet community (one of our projects at the Collaborative Knowledge Foundation) has built. Parts of the new submission system are still in development, but a slice of it is in production with hundreds of submissions going through it already: https://elifesciences.org/inside-elife/13f2ccb2/for-authors-a-simpler-way-to-submit-to-elife
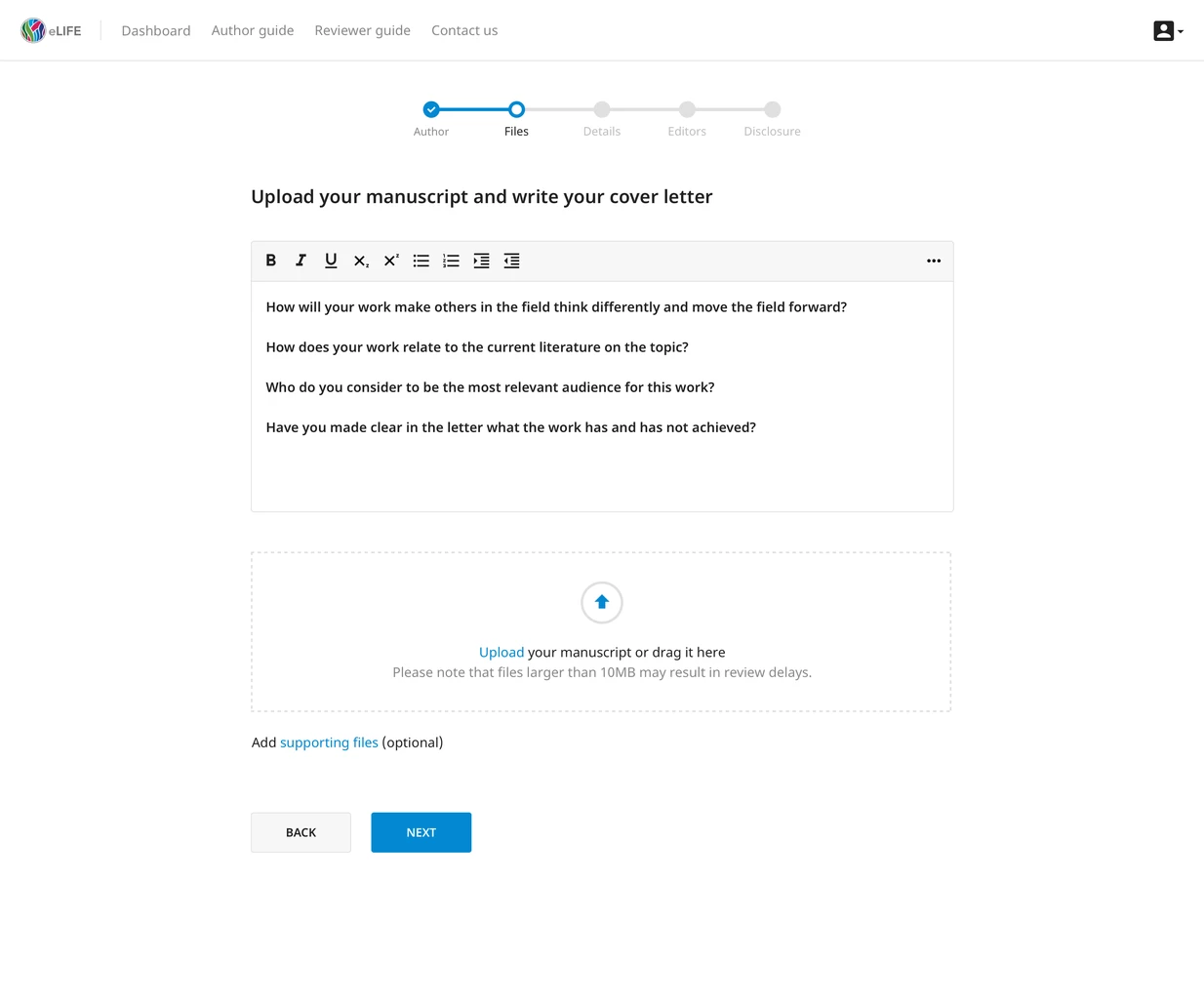
There are a lot of exceptional qualities to this thing, but there's three that stand out in my mind:
- eLife is using machine learning through its ScienceBeam project, to extract semantic data from submitted documents and make the process easy submitting authors. Goodbye endless forms where you have to retype everything, twice! As the volume of research globally goes up, machine learning will have an increasingly important role to play, especially for input help (as above) and quality control tasks (like checking statistics).
- The design and development of it are done completely in the open over at https://github.com/elifesciences/elife-xpub
- eLife is developing this system in collaboration with the PubSweet community (members include Hindawi, EBI, Micropublication, and others). I think it takes a lot of courage to join a group like that, share your plans and ideas with it, and face challenges together - that's commendable in its own right. But then, as has recently transpired, to have the open mind to evaluate and accept high-level software architecture changes proposed outside of your own organisation (Hindawi's architecture is making its way to eLife), that truly is exceptional. Perhaps there's a story here that should be told on its own, but there are surely great things coming from this project and collaboration in the future.
Onwards!
PREreview
We're talking about peer reviews for preprints. Makes sense, right? This one's development is just getting started as the project semi-recently secured some funding:
🚀We are so excited to announce that we have been awarded ✨two grants✨, one from @SloanFoundation and one from @wellcometrust in collaboration w @outbreaksci. This is a great opportunity for us to grow our community. Thank you all for your support! https://t.co/tirc7NH3PK
— PREreview (@PREreview_) December 7, 2018
Why would one limit what type of scientific content can get reviewed? Preprints are people too! In other words, PREreview makes post-publication reviews a reality on a massive scale, and that will have numerous beneficial side(and network)-effects. As Daniela Saderi, PREreview's co-founder very eloquently puts it:
“Our goal is to facilitate a cultural shift in which every scientist posts, reads, and engages with preprints as standard practice in scholarly publishing. We want to help scientists see the benefits of both sharing their work openly and exchanging timely feedback, in a way that is constructive, inclusive, and rewarding to them.”
👏
Development for this project is also in the open (notice a trend here?), you can follow along over here: https://github.com/PREreview/PREreview-2.
Micropublications 🔍
Another bias alert here, the folks behind https://www.micropublication.org/ are also part of the PubSweet community. I don't know, maybe it's just a cool community 🤷♂.️
The idea behind micropublications (micropubs for friends) is quite self-evident: publish small bits of research (a paragraph plus a figure or table), and do it quickly (the length of the publication process is measured in hours and days, not months). The system is being developed in the open as well, over here at https://gitlab.coko.foundation/micropubs/wormbase. Initially, the scope of the research published in this system will be C. elegans research, but plans to expand beyond that are already in motion.
The approach of publishing granular findings early and quickly is something that should find its way into many fields and systems out there.
Meta
This one's a bit different in that: 1. it tackles the reading side of the scientific publishing equation (if we consider submissions to be the 'writing side'), and 2. I haven't actually been able to try it out yet. There's a waiting list, it's closed source and things like that 🤷♂️. However, the approach they're taking, of using natural language processing (concept mining) and other machine learning solutions under the hood, with results presented in a user-friendly manner, is innovative and notable. Meta uses machine learning to guide you through discovering new relevant papers and exploring new concepts in your field.
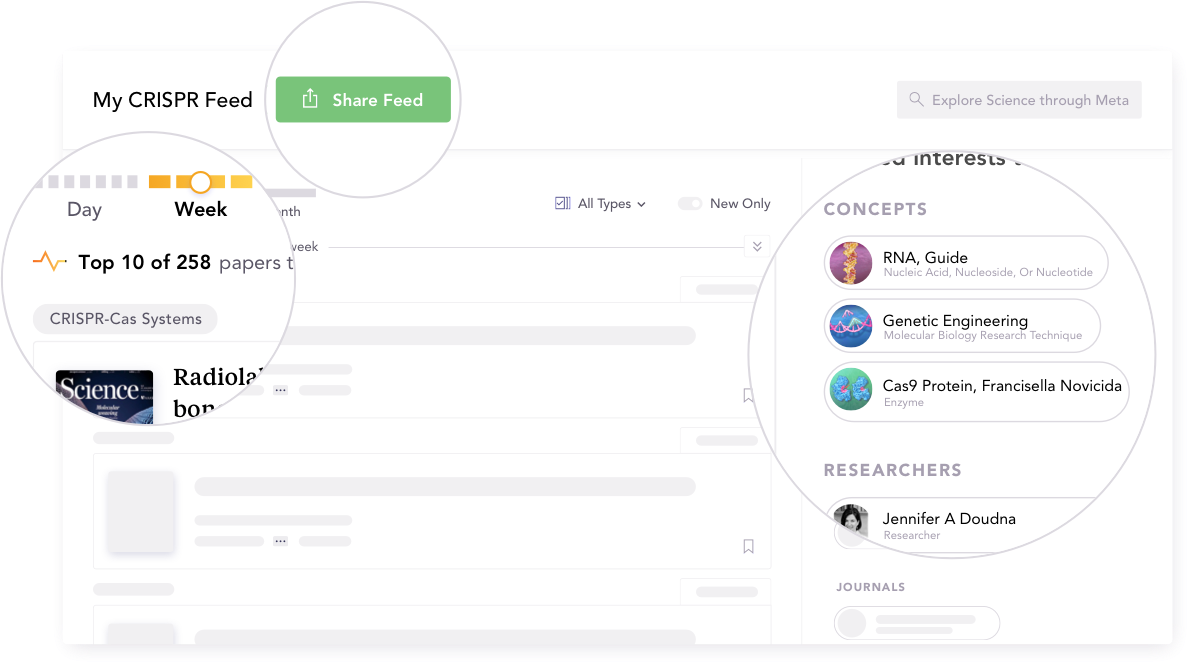
Researcher overload is a real problem, one that's only recently appeared, but also one that's unlikely to disappear anytime soon. With the rapidly increasing number of publications per unit of time (this includes preprints), solutions that help you not miss an important newly published result in a sea of irrelevant papers are sorely needed.
Again, I just wish Meta was open. At least some parts of it could be made open? 🙏
And that's the end of my list!
Update: Below are some great projects suggested by the good folks at Twitter.
- Stencila: An open source office suite for reproducible research.
Stencila was founded by my good friend Nokome Bentley (what's up Nokome!) and is a project with great potential. Yes, you've guessed it, it too is developed openly, you can follow along here: https://github.com/stencila and on their Twitter. I think they're currently focusing on some of the required under-the-hood things, likenixster(dependency management using Nix),dockter(building containers for research) and executable documents schema. - Dat project: A protocol for researcher-centric data sharing.
Sharing large datasets is difficult with current tech as you must, in almost all situations, rely on centralised storage, which can be expensive and time-consuming. Dat aims to solve this by providing an easy-to-use decentralised storage layer. One of the most interesting things that use Dat is Beaker, a browser and a content-production app in one application. One-click decentralised publishing, anyone? Yes, please! Follow along on their Twitter. - JOSS: The Journal of Open Source Software.
Exactly what it says on the tin, JOSS is an open access journal for open source software. It also runs on open source software (https://github.com/openjournals/joss) and has this wonderful slow web vibe to it. Here's an example of a publication about a package for microbiome analysis. Super minimal, fast and to the point. And here's their twitter. - Open Citations: Open bibliographic and citation data.
- Plaudit: A browser extension that enables verified (via ORCID) endorsements of publications. A way to let other researchers know what you think is good science.
- PubPub: Collaborative writing and publishing.
- ReScience: A journal for replicated computational research.
Lives on GitHub at https://github.com/ReScience/ReScience, and all replication results are published there. For example, a study that replicates a computational study of the conductance of pituitary cells in certain conditions. Reproducible science is good, but replicated science is better, they say. I agree! - FlashPub: Fast micropublications.
A platform for publishing very tiny bits of research very fast. It's in development, said to be launching in 2020, and one can sign up for the beta over here: https://flashpub.io/beta. I don't see a link to any code or any other content, so I assume the development is closed. - ReimagineReview: A whole database of peer review innovations!
This registry by ASAPbio contains more than 30 projects that aim to do things differently in the land of peer review, some of which we've already mentioned, but definitely check it out! - Dokieli: Decentralized articles and annotations, using linked data.
- Scite.ai: Machine learning for determining research quality.
This is an interesting approach, where natural language processing is used to look at papers that cite a given paper, and figure out if the given paper is supported or refuted by the citation. - Scholarcy: Automatic summarisation of papers.
Uses machine learning to provide a summary of a given paper in an effort to combat the 'too much info, too little time' problem that's spreading like wildfire (everywhere, but also research). - BMJ Labs' mega list: Projects in scientific publishing, all of them!
Alright folks, this is it. Everybody can go home now. This list links to not 100, not 200, but over 700 projects. At this point in time, it's 753, to be exact. This list makes me feel very humble and small, though there is immense strength in this incredible diversity. But perhaps it can also serve as a reminder, that we should also reach out and collaborate with other projects more often. There is also great strength in unity.
Keep the suggestions coming!
Your project here
I'm sure there are lots more cool things out there that I don't know about, so if you're working on an awesome project in this space let me know through the usual channels (me AT juretriglav.si or @juretriglav) and I'll include it here.
Thoughts, corrections, ideas, all of it is very welcome! Thanks for reading!
