Evolution of limb development in cephalopod mollusks
Oscar A Tarazona1,2, Davys H Lopez1, Leslie A Slota2, Martin J Cohn1,2*
1Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, United States; 2Department of Biology, UF Genetics Institute, University of Florida, Gainesville, United States
* For correspondence: mjcohn@ufl.edu
Abstract
Cephalopod mollusks evolved numerous anatomical novelties, including arms and tentacles, but little is known about the developmental mechanisms underlying cephalopod limb evolution. Here we show that all three axes of cuttlefish limbs are patterned by the same signaling networks that act in vertebrates and arthropods, although they evolved limbs independently. In cuttlefish limb buds, Hedgehog is expressed anteriorly. Posterior transplantation of Hedgehog-expressing cells induced mirror-image limb duplications. Bmp and Wnt signals, which establish dorsoventral polarity in vertebrate and arthropod limbs, are similarly polarized in cuttlefish. Inhibition of Bmp2/4 dorsally caused ectopic expression of Notum, which marks the ventral sucker field, and ectopic sucker development. Cuttlefish also show proximodistal regionalization of Hth, Exd, Dll, Dac, Sp8/9, and Wnt expression, which delineates arm and tentacle sucker fields. These results suggest that cephalopod limbs evolved by parallel activation of a genetic program for appendage development that was present in the bilaterian common ancestor.
https://doi.org/10.7554/eLife.43828.001eLife digest
Legs, wings, flippers and tentacles are just some examples of the diverse variety of animal limbs. Despite striking differences in form and function, all limbs develop in embryos using similar fundamental processes, like producing an outgrowth from the body and placing structures such as fingers, feathers, or suckers at appropriate positions. Animals have solved this problem multiple times during the history of life on Earth, in that limbed animals have arisen from limbless ancestors on many separate occasions. It is not clear, however, whether the same genetic instructions shape the developing limbs of all species.
Species that have limbs fall under three main groups of animals: arthropods, such as insects and crustaceans; vertebrates, like amphibians, reptiles and mammals; and a specialized group of mollusks known as cephalopods, which includes squid, cuttlefish and octopuses. It has been over two decades since the discovery that the limbs of vertebrates and insects develop using a similar molecular recipe, but the mechanisms responsible for the limbs of cephalopods had not been determined.
Tarazona et al. have now established that the genetic mechanisms that control how cuttlefish limbs develop are the same as those used by the limbs of vertebrates and insects. These mechanisms are also applied for similar purposes in each animal group. Notably, a signaling pathway called hedgehog, which controls the number of fingers that develop on a hand, also dictates the number of suckers on a cuttlefish arm. This may mean that an ancient system for creating limbs emerged over 500 million years ago in the earliest animals with bilateral symmetry (i.e., animals with mirror image halves), and activating this ancient genetic program resulted in the evolution of limbs in different animal lineages.
The extent of the genetic similarities between cuttlefish, mammals and insects suggests that this mechanism is likely to provide instructions about where cells position themselves in the developing limb. The next step is to examine how these common systems are interpreted differently to give arms, legs, wings and other limb forms.
https://doi.org/10.7554/eLife.43828.002Introduction
Animal appendages have widely varying morphologies and perform a multitude of functions, including locomotion, feeding, and reproduction (Nielsen, 2012; Ruppert et al., 2004). Limbs evolved on multiple occasions, and the absence of shared ontogenetic or morphological precursors of appendages in many animal lineages is consistent with their independent origins (Minelli, 2003; Pueyo and Couso, 2005; Shubin et al., 1997). This has led to the view that appendages in different clades of Bilateria are non-homologous morphological innovations that arose by convergent evolution (Nielsen, 2012; Ruppert et al., 2004). However, despite more than 500 million years of divergence, the independently evolved limbs of arthropods and vertebrates share developmental genetic similarities (Pueyo and Couso, 2005; Shubin et al., 1997; Tabin et al., 1999).
These discoveries led to debate over whether the genetic program for appendage development evolved in the common ancestor of all bilaterians in the early Cambrian, or whether arthropod and vertebrate appendages have undergone rampant convergence of developmental programs (Minelli, 2000; Minelli, 2003; Panganiban et al., 1997; Pueyo and Couso, 2005; Shubin et al., 1997; Tabin et al., 1999). A major obstacle to resolving this question is that the evidence of a conserved program derives almost exclusively from Ecdysozoa and Deuterostomia (Pueyo and Couso, 2005; Shubin et al., 1997), and little is known about molecular mechanisms of limb development in Spiralia, the third major superphylum of Bilateria (Grimmel et al., 2016; Prpic, 2008; Winchell and Jacobs, 2013; Winchell et al., 2010).
Within spiralians, the phylum Mollusca is the largest lineage, displaying a rich diversity of body plans (Figure 1A) dating back to the Cambrian explosion (Ruppert et al., 2004; Smith et al., 2011). The evolution of arms and tentacles in cephalopod mollusks contributed to the successful adaptive radiation of these agile marine predators (Kröger et al., 2011; Ruppert et al., 2004). Cephalopod limbs are highly muscular appendages that bear cup-shaped suckers on their ventral sides. Arms are short and have suckers along the entire ventral surface (Figure 1B and C), whereas tentacles are longer, retractable appendages with suckers restricted to a distal pad (Figure 1D and E). Tentacles are thought to be specialized serial homologs of the arms (Arnold, 1965; Lemaire, 1970; Shigeno et al., 2008) and are present in decapods (squid and cuttlefish) but absent in nautilids and octopods. Limbs likely evolved de novo in cephalopods (Figure 1A), since no homologous precursor structures have been identified in any other mollusk lineages (Lee et al., 2003; Shigeno et al., 2008). To test the hypothesis that cephalopod limbs evolved by recruitment of an ancient gene regulatory network for appendage development that is conserved across Bilateria, we investigated arm and tentacle development in embryos of the cuttlefish, Sepia officinalis.
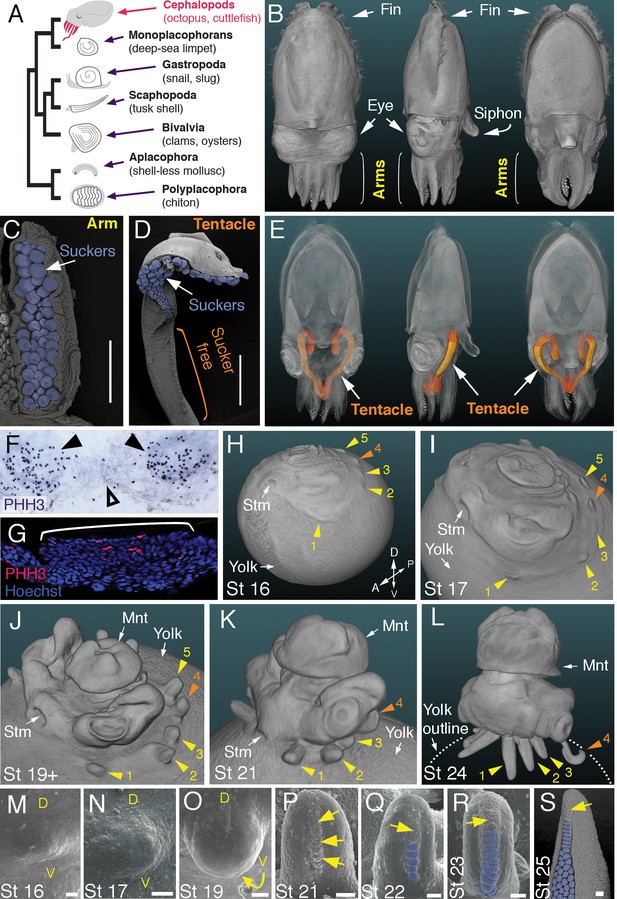
Development of arms and tentacles in the cuttlefish, Sepia officinalis.
(A) Phylogenetic relationships of Mollusca based on phylogenomic data (Smith et al., 2011) illustrating the unique morphology of the cephalopod body plan compared to other mollusks. (B) OPT reconstruction of a cuttlefish hatchling showing positions of the limbs; only arms are visible (see also Figure 1—video 1). (C to D), SEM of the ventral side of a cuttlefish arm (C) and tentacle (D). Suckers are pseudocolored blue. Note distal restriction of suckers in tentacle relative to arm. (E) OPT reconstruction illustrating the internally retracted tentacles. Specimens are same as in (B), but here the tentacles are displayed in orange and the rest of the tissue is rendered translucent (see also Figure 1—video 2). (F and G) Phospho-histone H3 (PHH3) immunostaining at stage 15 shows localized clusters of proliferating cells at the onset of limb development (black arrowheads) but little proliferation in the interlimb region (open arrowhead). (F) PHH3 detection by colorimetric reaction with DAB in a whole mount. (G) PHH3 immunofluorescence (red) on limb cryosection (white bracket). Cell nuclei (blue) are labeled by Hoechst staining. (H-L) OPT reconstructions of cuttlefish embryos at stages 16 to 24. Cuttlefishes have five bilaterally symmetric limb pairs (ten limbs; eighth arms and two tentacles). Numbered arrowheads mark all five limbs/limb buds on the left side of each embryo. The left tentacle differentiates from position number four (orange arrowhead), whereas arms form from limb buds at the other positions (yellow arrowheads). See also Figure 1—videos 3—5. A, anterior; P, posterior; D, dorsal; V, ventral; Stm, stomodeum; Mnt, mantle. (M to O) SEM during early stages of cuttlefish limb development (stages 16 to 19). Morphogenesis of the limb is first observed as a slight swelling (M) that transforms into a limb bud (O) as proximodistal outgrowth progresses. D, dorsal; V, ventral. (P–S), SEM at later stages of cuttlefish limb development (stages 21 to 25) showing the formation of sucker buds on the ventral surface of a developing limb. A primordial sucker band (yellow arrows) is observed along the ventral midline of a stage 21 limb bud (P). At later stages, the band cleaves superficially from the proximal end to form the sucker buds (pseudocolored blue in Q to S). Scale bars: 0.5 mm (C and D) and 100 μm (M to S).
Results
Development of arms and tentacles in the cuttlefish (Sepia officinalis)
Cuttlefishes are decapod cephalopods that have eight arms and two tentacles (Figure 1B–E; Figure 1—videos 1 and 2). Fertilized cuttlefish eggs undergo superficial cleavage, and scanning electron microscopy and optical projection tomography show that most embryonic development is restricted to the animal pole (Figure 1H and I). The first sign of limb formation is observed at stage 16, when all ten limb primordia (five on each side) can be detected as small swellings around the periphery of a flat-shaped embryo, which lies at the top of the large yolk mass (Figure 1H and M). Analysis of the mitotic marker phospho-histone H3 (PHH3) at stage 15 revealed localized clusters of PHH3-positive cells in each of the early limb primordia (Figure 1F and G), indicating that initiation of limb outgrowth is caused by localized cell proliferation. Discrete limb buds are observed from stage 17 (Figure 1I and N; Figure 1—video 3). As the embryo begins to rise-up on the animal pole around stage 19, the limb buds start to elongate along the proximodistal axis (Figure 1J and O; Figure 1—video 4), and by stage 24, the differential length and morphology of arms relative to tentacles is apparent (Figure 1L; Figure 1—video 5).
Analysis of sucker development showed that a sucker field primordium initially forms as a narrow proximodistal ridge along the ventral surface of each limb (evident by stage 21; Figure 1P). At later stages, the sucker field ridge cleaves superficially, segregating sucker buds from proximal to distal (Figure 1Q). As the arms elongate, the sucker buds are laid down on the entire ventral surface of each arm (Figure 1L and R; Figure 1—figure supplement 1A and C–G), forming four parallel rows across the anteroposterior axis (Figure 1C; Figure 1—figure supplement 1A). In the tentacles, the primordial sucker band is restricted to the distal tip, where sucker buds form in eight rows along the anteroposterior axis of the tentacle sucker pads (Figure 1D; Figure 1—figure supplement 1B). The full complement of immature sucker bud rows is present on each limb at hatching, and differentiation of the suckers continues during post-hatch development (Figure 1—figure supplement 1H and I).
Molecular analysis of cuttlefish limbs reveals conservation of proximodistal, anteroposterior, and dorsoventral patterning networks
To test the hypothesis that cuttlefish limb development is regulated by the same molecular mechanisms that pattern arthropod and vertebrate limbs, despite their independent evolutionary origins, we cloned and characterized cuttlefish orthologs of genes that pattern the three axes of vertebrate and arthropod limbs, and then analyzed their expression patterns during cuttlefish limb development (Figure 2 and Figure 2—figure supplements 1–10).
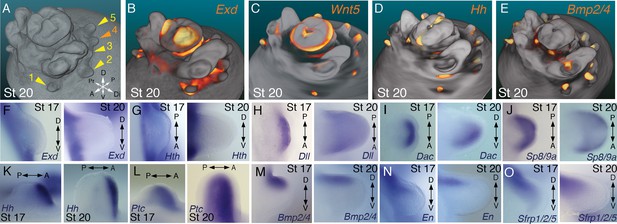
Molecular regionalization of proximodistal, anteroposterior, and dorsovental axes during cephalopod limb development.
(A) OPT reconstruction of cuttlefish embryo at stage 20 showing all five limb buds on the left side of the embryo (arms, yellow arrowheads; tentacle, orange arrowhead). (B to E) OPT reconstructions showing four representative genes with polarized expression patterns along major axes of limb buds (gene expression indicated by orange/yellow). Proximodistally polarized expression of Exd (B) and Wnt5 (C). Anteroposteriorly polarized expression of Hh (D), dorsoventrally polarized expression of Bmp2/4 (E). (F to O), In situ hybridizations of cuttlefish limb buds at stage 17 (left) and stage 20 (right) showing polarized patterns of expression along the proximodistal axis for Exd (F), Hth (G), Dll (H), Dac (I) and Sp8/9a (J); the anteroposterior axis for Hh (K) and Ptc (L); and the dorsoventral axis for Bmp2/4 (M), En (N) and Sfrp1/2/5 (O). A, anterior; P, posterior; D, dorsal; V, ventral; Di, distal; Pr, proximal.
Partial sequences of cuttlefish cDNAs (Sepia officinalis and Sepia bandensis) were isolated by rt-PCR, and preliminary identities were determined by comparison with NCBI sequence databases, including the octopus genome. Molecular phylogenetic reconstructions were then made by maximum likelihood phylogenetic inference using the best amino acid substitution model for each gene family (see Materials and methods for details). Tree topologies with well-supported bootstrap values showed the position of each cuttlefish gene within the targeted gene families, which included Wnt, Tcf/Lef, Frizzled (Fzd), Dachsund (Dac/Dach), Notum, Patched (Ptc/Ptch), Hedgehog (Hh), Bone morphogenetic protein (Bmp), Specificity protein (Sp), and the ANTP and TALE homeobox gene families (trees are shown in Figure 2—figure supplements 1–10 and are described below; gene accession ID numbers and the data set used in the phylogenetic analyses is provided in Figure 2—source data 1).
Within the Wnt family of cell signaling proteins, we isolated cuttlefish orthologs of Wnt1, Wnt2, Wnt5, and Wnt7 (Figure 2—figure supplement 1). Phylogenetic analysis of cuttlefish transcription factors identified Tcf3/4, an ortholog of arthropod Pangolin and a pro-ortholog of vertebrate Tcf3 and Tcf4 (Figure 2—figure supplement 2), Dac, a pro-ortholog of vertebrate Dach1 and Dach2 (Figure 2—figure supplement 3), and Sp8/9, a pro-ortholog of vertebrate Sp8 and Sp9 (Figure 2—figure supplement 4). We also identified numerous homeobox genes, which phylogenetic analyses confirmed to be Dll, a pro-ortholog of vertebrate Dlx genes, Exd, a pro-ortholog of vertebrate Pbx genes, Hth, a pro-ortholog of vertebrate Meis1 and Meis2, and Engrailed, a pro-ortholog of vertebrate En1 and En2 (Figure 2—figure supplement 5).
In addition, we cloned the Wnt extracellular inhibitors Notum and Sfrp-1/2/5, and the Wnt co-receptor Fzd9/10 (Figure 2—figure supplements 6 and 7). Cuttlefish possess a Bmp-2/4 gene that is an ortholog of arthropod Dpp and a pro-ortholog of vertebrate Bmp2 and Bmp4 (Figure 2—figure supplement 8), a Hh gene (Grimaldi et al., 2008) that we show to be a pro-ortholog of the vertebrate hedgehog family (Figure 2—figure supplement 9), and a gene encoding the Hh receptor Patched, a pro-ortholog of vertebrate Ptch1 and Ptch2 (Figure 2—figure supplement 10). The cuttlefish Sfrp ortholog that we identified as Sfrp1/2/5 was annotated incorrectly in the octopus genome as Frizzled1 (Figure 2—source data 1). We also found two Sp8/9 genes in the octopus genome (Figure 2—source data 1), and the cuttlefish Sp8/9 gene shows clear orthology to only one of the two octopus genes (Figure 2—figure supplement 4), suggesting that the Sp8/9 gene underwent a duplication in cephalopod mollusks. Therefore, we designate the octopus Sp8/9 paralogs as Sp8/9a and Sp8/9b, and the cuttlefish Sp8/9 gene that we isolated is the ortholog of Sp8/9a.
We next investigated the spatial and temporal expression patterns of these genes during cuttlefish limb development. Genes that pattern the proximodistal axis of arthropod and vertebrate limbs (Lecuit and Cohen, 1997; Mercader et al., 1999; Panganiban et al., 1997; Pueyo and Couso, 2005) showed similarly polarized patterns of expression along the proximodistal axis of cuttlefish limb buds, with Exd and Hth restricted proximally (Figure 2B,F and G; Figure 3A–E; Figure 3—figure supplement 1A and B; and Figure 3—figure supplement 2A,I and J) and Dll, Dac, Sp8/9a, Wnt1, Wnt5, and Wnt7 restricted distally (Figure 2C,H–J; Figure 3F–I; Figure 3—figure supplement 1C–E and L–N; and Figure 3—figure supplement 2B,C,G,I and J). At stages 20–21, the distal expression boundaries of Exd and Hth and the proximal expression boundaries of Dll and Sp8/9a appear to mark the morphological boundary between the proximal sucker-free and the distal sucker-forming regions (compare right panels in Figure 2F–H and J with Figure 1P). Indeed, at stages when arms and tentacles begin to develop their distinctive morphologies -- tentacles are longer and have an extensive proximal sucker-free domain -- the Exd/Hth expression domains were found to extend further distally in tentacles (Figure 3B,D) compared to arms (Figure 3A and C). This distal expansion of the Exd/Hth expression domain matches the expanded sucker-free region and the distal restriction of suckers in tentacles (Figure 3E).
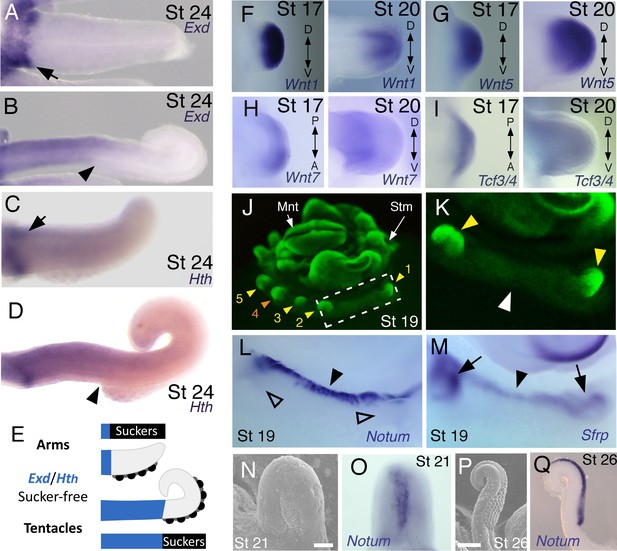
Expression of proximal identity genes Exd and Hth in arms and tentacles corresponds with distribution of suckers; Wnt signaling repressors are dorsally restricted.
(A and B) Compared to arms (A), tentacles (B) show a distally expanded domain of Exd expression in the proximal region of the limb. (C and D) A similar pattern of expression is detected for Hth during arm (C) and tentacle (D) development. Distal boundary of Exd and Hth expression marked by black arrowheads in (A to D). (E) Expanded expression of proximal identity genes correlates with the expanded sucker-free domain seen in tentacles compared to arms. (F and H) The Wnt ligands Wnt1, Wnt5 and Wnt7 show a distally restricted expression but no dorsoventral polarization at stages 17 and 20. (I) The Wnt signaling transcription factor Tcf3/4 is also distally restricted but shows no dorsoventral polarization at stages 17 and 20. (J and K) Fluorescent nuclear stain SYBR Safe highlights limb buds (yellow arrowheads). Boxed region in (J) is enlarged in (K); white arrowhead marks interlimb region. (L and M) The Wnt ligand repressors Notum and Sfrp1/2/5 are expressed in the dorsal interlimb region (black arrowhead in L and M; compare with K). Sfrp1/2/5 expression expands into the dorsal limb bud (black arrows in M) in stage 19 embryos, whereas Notum stays dorsal but proximally restricted (open arrowheads mark the limb buds in L). (N and O) The earliest sign of sucker formation can be detected by SEM as a slight swelling (N) and by Notum expression (O) on the ventral side of stage 21 limb buds. (P and Q) Expression of Notum is maintained through later stages of sucker morphogenesis, as seen in stage 26 tentacles (lateral views).
Our finding that the proximodistal axis of cuttlefish limbs shares patterns of molecular regionalization with arthropod and vertebrate limbs led us to examine whether anteroposterior and dorsoventral axis development are also conserved. Posteriorly polarized activation of Hedgehog signaling in arthropod and vertebrate limbs is essential for proper patterning of the anteroposterior axis, and ectopic activation of the Hedgehog pathway induces anterior duplication of posterior structures (Basler and Struhl, 1994; Kojima et al., 1994; Riddle et al., 1993). We analyzed Hh expression during cuttlefish limb development at stages 16 to 20 and found that Hh expression is also polarized to one side of cuttlefish limb buds. In cuttlefishes, however, Hh expression is restricted to the anterior margin of the limb bud, whereas in arthropods and vertebrates, Hh/Shh is expressed posteriorly (Figure 2D and K; and Figure 3—figure supplement 2D). Consistent with the anterior localization of Hh, we detected expression of Patched, which serves as a readout of Hedgehog signal transduction, in an anterior-to-posterior gradient (Figure 2L). Thus, anteroposteriorly restricted activation of the Hedgehog pathway is a conserved feature of cephalopod, arthropod, and vertebrate limb development, but the polarity of the signaling center is reversed in cephalopod limbs. By stage 21, the anteriorly restricted Hh domain has diminished and a new, central expression domain appears in the location of the brachial nerve primordia (Figure 3—figure supplement 1F,K).
We then examined the dorsoventral axis, which is controlled by the antagonistic actions of wg/Wnt and dpp/Bmp signaling in arthropods and vertebrates (Brook and Cohen, 1996; Cygan et al., 1997; Diaz-Benjumea et al., 1994; Jiang and Struhl, 1996; Parr and McMahon, 1995). In arthropods, the Wnt ligand wg is expressed ventrally, whereas the Bmp2/4 ortholog dpp is expressed dorsally (Basler and Struhl, 1994; Diaz-Benjumea et al., 1994). Expression and function of the Wnt-Bmp network is conserved, albeit with inverted polarity, in vertebrate limbs; Wnt7a is expressed dorsally (Parr and McMahon, 1995) and Bmp signaling activates Engrailed1 (En1) ventrally (Ahn et al., 2001), and these interactions regulate development of dorsal and ventral limb structures (Cygan et al., 1997; Parr and McMahon, 1995). During cuttlefish limb development, Bmp2/4 and En show dorsally polarized expression (Figure 2E,M and N; and Figure 3—figure supplement 2E). Genes encoding Wnt ligands (Wnt1, Wnt5 and Wnt7) and cellular components of canonical Wnt signaling cascade (Tcf3/4 and Frz9/10) are expressed broadly throughout the dorsoventral axis of cuttlefish limb buds (Figure 3F–I and Figure 3—figure supplement 1L–R; and Figure 3—figure supplement 2G,I and J); however, the secreted Wnt antagonists Notum and Sfrp1/2/5 are expressed dorsally in the limb and interlimb regions (Figure 3J–M), with the Sfrp1/2/5 domain extending deeper into the dorsal limb buds (Figure 2O; Figure 3M). This dorsal expression of Wnt antagonists suggests a mechanism for restriction of Wnt signaling to the ventral side of the cephalopod limb buds. Taken together, these results suggest that the genetic pathways active along the proximodistal, anteroposterior, and dorsoventral axes of cephalopod limbs are homologous (specifically, orthologous) to the networks that regulate limb development in arthropods and vertebrates.
In order to further test this hypothesis, we next performed a series of functional experiments to determine whether polarized expression of these signaling molecules is involved in patterning the anteroposterior and dorsoventral axes of cuttlefish limbs (described below). We developed a method for ex-ovo culture of cuttlefish embryos (see Material and methods) to allow in vivo manipulations of genetic pathways in early limb buds.
Bmp signaling controls dorsoventral patterning of cuttlefish limbs
A hallmark of dorsoventral polarity is the restriction of sucker buds to the ventral surface of the limb (Figure 1C,D and S), and this is preceded by ventral expression of Notum in the sucker-forming region at stage 21 (Figure 3N–Q). We asked whether polarized expression of Bmp2/4 on the dorsal side of cuttlefish limb buds is required for the specification of dorsal identity. To repress dorsal Bmp activity, we implanted carrier beads loaded with Noggin (Nog), a secreted Bmp inhibitor protein, on the dorsal side of stage 17 limb buds (Figure 4A). Implantation of Nog beads on the dorsal side of cuttlefish limb buds resulted in ectopic, dorsal expansion of the Notum mRNA domain (n = 3/3; control PBS [phosphate buffered saline] beads had no effect on Notum expression [n = 3/3]) (Figure 4G,H). To determine whether inhibition of dorsal Bmp signaling respecifies dorsal cells to form ventral structures, we repeated the experiment and allowed embryos to develop to stage 26–27. Analysis of limb morphology by scanning electron microscopy revealed the presence of ectopic sucker buds on the dorsal surface of Nog-treated limbs (n = 8/12; Figure 4B; Figure 4—figure supplement 1A and B). The ectopic dorsal suckers extended around the distal tip of the limb and joined the ventral sucker field. By contrast, in limbs that received control PBS beads dorsally, sucker buds were restricted to ventral surface and terminated at the normal dorsal-ventral boundary at the tip of the limb (n = 15/15; Figure 4C). Our finding that antagonism of Bmp signaling results in development of ventral structures (sucker buds) on the dorsal side of the limb indicates that dorsal Bmp2/4 activity is required for the early specification of dorsal identity in cephalopod limb development.
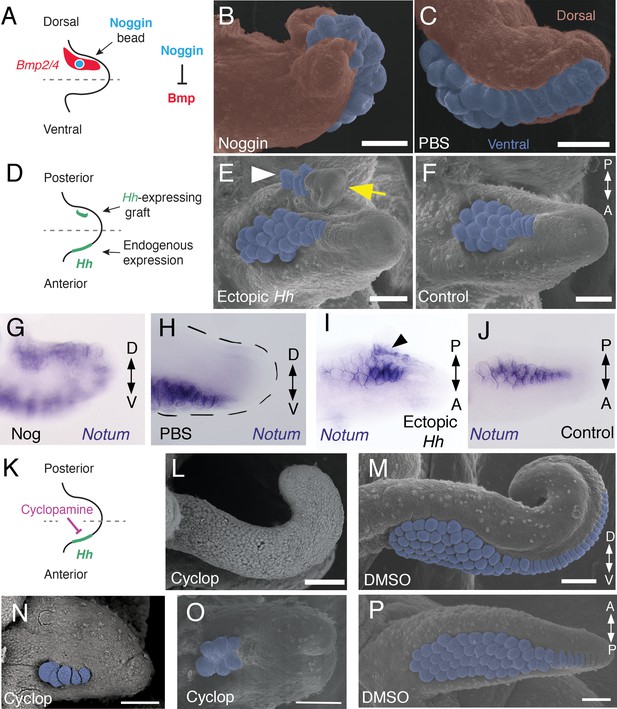
Bmp signaling controls dorsoventral patterning and Hh signaling regulates anteroposterior patterning of cuttlefish limbs.
(A to C) Implantation of carrier beads loaded with the Bmp inhibitor Noggin (A) results in formation of ectopic sucker buds (n = 8/12) on the dorsal surface of the limb (B), whereas PBS control beads (n = 15/15) result in normal development of the dorsal limb (pseudocolored red) restriction of suckers (pseudocolored blue) to the vental side of the limb (C). (D-F), Hh-expressing cells (taken from the funnel of a stage 24 donor embryo) grafted to the posterior side of a stage 17 cuttlefish limb bud (D) generates a posterior mirror-image limb duplication (n = 7/12; yellow arrow in E), whereas no duplication (n = 8/8) results when control (Hh-negative) cells are grafted to the same position (F). Sucker buds are pseudocolored blue in (E and F); sucker buds in duplicated limb marked with a white arrowhead. (G and H) Noggin beads induce ectopic expression of Notum on the dorsal side of the limb (G). Limbs receiving control PBS beads show normal expression of Notum ventrally (H). (I and J) Graft of Hh-expressing tissue to the posterior side of the limb induces ectopic domain of Notum prior to duplication of the limb (I; black arrowhead). Note the two separate domains of Notum expression in I compared to a single Notum expression domain in the limb with the Hh-negative control graft (J). (K to P) Transitory repression of Hh signaling by cyclopamine (K) during early stages of limb development disrupts the anteroposterior distribution of sucker bud rows (L, N, O). Cyclopamine-treated limbs showing complete loss of suckers in tentacles (L) and reduction in the number of sucker bud rows in arms (N and O). Control embryos treated with vehicle only (DMSO) develop the normal number of sucker bud rows in tentacles (M) and arms (P). Sucker buds are pseudocolored blue in (B, E, F, and M- P). Scale bars 100 μm.
Hedgehog signaling at the anterior margin of cuttlefish limb buds controls anteroposterior patterning of the sucker field
We then investigated whether the mechanism of anteroposterior patterning is conserved between cephalopod and vertebrate/arthropod limbs. To determine whether the anterior expression of Hh in cuttlefish limb buds controls anteroposterior patterning, we grafted Hh-expressing cells from the thickened funnel epithelium (Tarazona et al., 2016) to the posterior side of stage 17 limb buds, which created an ectopic source of Hh opposite the endogenous Hh expression domain (Figure 4D). We used Hh-expressing cells from the funnel, rather than the anterior side of the limb bud, to exclude the possibility of grafted limb cells undergoing self-differentiation. Transplantation of Hh-expressing cells to the posterior side of cuttlefish limb buds resulted in posterior limb duplications (n = 7/12; Figure 4E and Figure 4—figure supplement 1C,D). Analysis of morphology and gene expression in host limbs approximately 10 days after receiving the graft revealed that the posterior duplications even contained sucker buds, which were marked by Notum expression (Figure 4I and J). By contrast, limbs that received control grafts of stage 24 funnel epithelium that lacks Hh expression (Tarazona et al., 2016) developed normally (n = 8/8; Figure 4F).
Although these results suggest that Hh is sufficient to re-specify anteroposterior polarity in cuttlefish limbs, we wanted to exclude the possibility that posterior identity was induced by other factors that could be present in the graft. Therefore, we tested whether Hh signaling is necessary for anteroposterior patterning of cephalopod limbs by specifically repressing endogenous Hh signaling. A notable morphological feature of cephalopod limbs is the anteroposterior arrangement of parallel sucker rows on the ventral surface (Figure 1C,D and S). Based on the results of the transplantation experiments, we reasoned that Hh signaling could regulate the number of sucker rows along the anteroposterior axis of cephalopod limbs, similar to the manner in which Hh specifies digit number along the anteroposterior axis of vertebrate limbs (Lewis et al., 2001; Scherz et al., 2007; Zhu et al., 2008).
Transitory treatment (2 days) of cuttlefish embryos at stage 16, when Hh is first expressed on the anterior side of the early limb bud, with the small molecule cyclopamine, an inhibitor of Smoothened that represses Hh signaling (Figure 4K), disrupted the anteroposterior distribution of sucker rows in arms and tentacles. Severity of this phenotype ranged from arms with a reduced number of suckers and sucker rows (n = 10/10; Figure 4N and O) to completely sucker-free tentacles (n = 8/10; Figure 4L). Control treatments with vehicle only (DMSO) did not alter the normal anteroposterior pattern of sucker rows (n = 8/8; Figure 4M and P). Finally, to confirm that the phenotype of cyclopamine-treated embryos was not due to failure in brachial nerve differentiation, we examined acetylated tubulin immunofluorescence, which shows that the brachial nerve cords develop in both cyclopamine and DMSO treated embryos (Figure 4—figure supplement 1E,F). These results show that Hh signaling is necessary for proper patterning of the anteroposterior axis in cephalopod limb development.
Discussion
Our finding that the proximodistal, dorsoventral, and anteroposterior axes of cuttlefish limb buds are patterned by the same pathways that regulate arthropod and vertebrate limb development suggests that the independent evolution of limbs in cephalopod mollusks involved recruitment of an ancient genetic program for appendage development. Discovery of this appendage developmental circuit within Spiralia demonstrates its deep conservation across all three branches of Bilateria (i.e., Deuterostomia, Ecdysozoa, and Spiralia), suggesting its presence in the common ancestor of all bilaterians (Figure 5). Parallel recruitment of this ancient developmental genetic program may have played a role in the independent evolution of a wide diversity of appendages in many bilaterian lineages (Moczek and Nagy, 2005; Shubin et al., 2009).
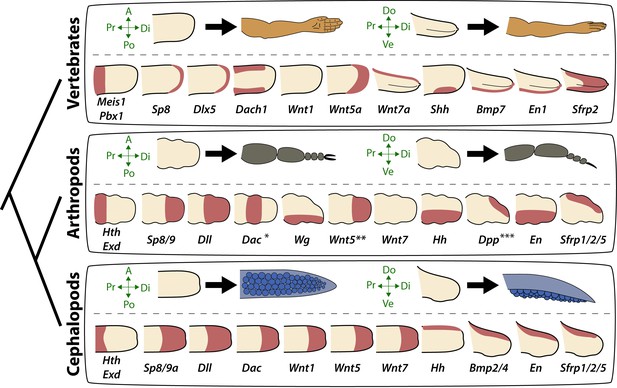
Molecular patterning of the anteroposterior, proximodistal, and dorsoventral axes of developing limbs in vertebrates, arthropods, and cephalopods.
For each lineage, the top row shows schematic representations of a generalized limb bud and an adult limb in two different orientations. Axes are indicated to the left of each limb bud (A, anterior; Po, posterior; Pr, proximal; Di, distal; Do, dorsal; Ve, ventral) Bottom rows show limb buds with gene expression domains pink color). Vertebrate gene expression based on mouse limb development. Arthropod gene expression is a compound reconstruction from chelicerate, myriapod, and hexapod limb development in order to consolidate a complete set of pro-orthologous genes comparable to vertebrate and cephalopod lineages. Cephalopod gene expression is based on findings in this study from the cuttlefish Sepia officinalis and Sepia bandensis. The figure illustrates the conserved and divergent expression patterns of homologous (orthologous) genes, some of which share equivalent roles in patterning the limb axes. The proximodistal axis displays conserved expression of transcription factors at opposite ends; Hth (pro-ortholog of vertebrate Meis genes) and Exd (pro-ortholog of vertebrate Pbx genes) are restricted proximally, whereas Dll (pro-ortholog of vertebrate Dlx genes), Wnt5 (pro-ortholog of Wnt5a) and Sp8/9 (pro-ortholog of vertebrate Sp8 and Sp9 genes, known as Sp6-9 in some arthropods) show distally restricted expression. The typical expression pattern of Dac seen in arthropods (between proximally and distally restricted genes) is not strictly conserved in vertebrates (Dac is the pro-ortholog of vertebrate Dach genes) or cephalopods. However, Dac expression in non-locomotory arthropod appendages (e.g., mandibles) is distally restricted, resembling cephalopod Dac expression (Donoughe and Extavour, 2016). Expression patterns of the diverse family of Wnt genes shows interesting variation. Although, some members of the family show variation in their expression pattern (Wnt1 and Wnt7), there is a general pattern of distal restriction of Wnt expression (represented here by Wnt5, but also seen in many other Wnt ligands) in the three lineages. At the level of individual Wnt members, Wg (pro-ortholog of vertebrate Wnt1) is restricted ventrally in arthropods but not in vertebrates or cephalopods, and Wnt7a (arthropod and cephalopod Wnt7 genes are pro-orthologs of vertebrate Wnt7a) is restricted dorsally in vertebrates but not in arthropods or cephalopods. Restricted expression of Wnt ligands either dorsally or ventrally has not been reported in cephalopods, but the dorsally restricted expression of the Wnt repressor Sfrp1/2/5 suggests a role of polarized Wnt pathway activation in the control of the dorsoventral axis of cephalopod limbs, similar to vertebrates (by dorsal Wnt7a) and arthropods (by ventral Wg). There is a clear restriction of at least one Bmp ligand (vertebrate Bmp7 and cephalopod Bmp2/4; pro-orthologs of arthropod Dpp) and the transcription factor En along the dorsoventral axis in these three lineages. Finally, polarized expression of Hh is conserved in the three lineages (posterior in vertebrates and arthropods, but anterior in cephalopods), which, together with the functional manipulations, indicates conservation of Hh signaling in patterning the anteroposterior limb axis in the three lineages. The asterisk (*) in arthropod Dac indicates that some mouth appendages show a distal expression domain (Donoughe and Extavour, 2016) (Angelini and Kaufman, 2005) more similar to cephalopod Dac limb expression than to Dac expression in arthropod legs. Two asterisks (**) indicate that Wnt5 expression shows variation in arthropods, with a sub-distal expression in chelicerates (Damen, 2002) but distal in hexapods (i.e. flour beetle) (Bolognesi et al., 2008). Three asterisks (***) indicate that Dpp shows variation in its expression domain in arthropods, with some hexapods and chelicerates showing a distal expression domain, whereas in Myriapods and other hexapods it is dorsally restricted, as depicted here (Angelini and Kaufman, 2005). Schematized gene expression domains for vertebrates and arthropods are from the following sources. Mouse gene expression: Meis1 (González-Lázaro et al., 2014), Pbx1 (Capellini et al., 2006), Sp8 (Kawakami et al., 2004), Dlx5 (Vieux-Rochas et al., 2013), Dach1 (Salsi et al., 2008), Wnt1, Wnt5a, Wnt7a, Sfrp2 (Witte et al., 2009), Shh (Riddle et al., 1993), Bmp7 (Choi et al., 2012) and En1 (Loomis et al., 1998). Arthropod expression based on: Chelicerates, Hth, Exd, Dll, Dac (Prpic et al., 2003), Sp8/9 (Königsmann et al., 2017), Wg (Damen, 2002), En (Damen, 2002) and Sfrp1/2/5 (Hogvall et al., 2018); Myriapods, Hth, Exd, Dll, Dac (Prpic and Tautz, 2003), Sp8/9 (Setton and Sharma, 2018), Wg, Dpp (Prpic, 2004), Wnt5, Wnt7, Hh, En (Janssen et al., 2004), Sfrp1/2/5 (Hogvall et al., 2018); Hexapods, flour beetle, Hth, Exd (Prpic et al., 2003), Sp8/9 (Schaeper et al., 2010), Dll (Beermann et al., 2001), Dac (Prpic et al., 2001), Wg, Wnt5 (Bolognesi et al., 2008), Dpp (Sanchez-Salazar et al., 1996), En (Brown et al., 1994); Hexapods, cricket, Hth, Exd, Dll, Dac, Wg, Hh, Dpp, En (Donoughe and Extavour, 2016).
The discovery that cephalopod, arthropod, and vertebrate appendages develop using conserved developmental mechanisms does not exclude the possibility that other types of appendages evolved by recruiting a different set of developmental tools (or by utilizing the same tools but in different patterns). Examination of gene expression in lateral parapodial appendages of the polychaete worm Neanthes, also a spiralian, led to the suggestion that the molecular mechanisms of polychaete appendage development might not be conserved with ecdysozoans and deuterostomes (Winchell and Jacobs, 2013; Winchell et al., 2010). However, given that relatively few genes were examined in Neanthes parapodia, it is difficult to conclude whether the reported differences between parapodia and arthropod/vertebrate/cephalopod limbs reflect the unique nature of parapodia or lineage-specific divergences that occurred after recruitment of the core developmental program. A study of a different polychaete, Platynereis dumerilii, showed that gene expression is generally conserved in appendages that form during regeneration of caudal trunk segments, although some divergent patterns were observed and these were suggested to reflect taxon-specific differences in appendage morphology (Grimmel et al., 2016). How parapodia fit into the picture of animal appendage evolution will require additional studies of spiralian appendages to increase the diversity of species, types of appendages, and number of genes/pathways interrogated. Nonetheless, our discovery that cephalopod arms and tentacles evolved by parallel recruitment of the same genetic program that orchestrates appendage formation in arthropods and vertebrates suggests that this program was present in the bilaterian common ancestor.
Activation of this ancient developmental program could also underlie the origin of other morphological innovations, including non-locomotory appendages such as beetle horns (Moczek and Nagy, 2005; Moczek et al., 2006) and external genital organs of amniote vertebrates (Cohn, 2011; Gredler et al., 2014). We propose that the genetic program for appendage formation was stabilized in Bilateria, including those lineages that lack limbs, for development of appendage-like structures. This hypothesis implies that the ancestral appendage developmental program was not a latent developmental feature that was redeployed each time that limbs evolved, but rather it might have been a continuously activated network that controlled formation of outgrowths in general.
One of our observations raises the possibility that the gene network that controls appendage formation could be conserved in non-cephalopod mollusks, despite the absence of arms and tentacles in those lineages. During cuttlefish funnel/siphon development, we found asymmetric expression of Hh (Tarazona et al., 2016) and proximodistally polarized expression of Wnt5 and Exd, which partially mirror their expression patterns during arm and tentacle development (Figure 5—figure supplement 1). If this gene network is found to be active in the developing funnel/siphon of non-cephalopod mollusks, then the funnel/siphon would represent a more primitive site of expression in mollusks, given that evolution of the molluscan funnel/siphon predates the origin of cephalopod limbs (Nielsen, 2012; Ruppert et al., 2004). Further studies of gene expression and function during funnel/siphon development in mollusks will be needed to determine if this clade shows conservation of the appendage development program beyond cephalopod arm and tentacle development.
Although the bilaterian common ancestor may have used this genetic program to control development of rudimentary outgrowths (e.g., appendages, funnel/siphon, genitalia), it is also possible that it predates the evolution of locomotory and non- locomotory appendages. Studies of cephalic neuroectoderm showed that gene expression patterns controlling the anteroposterior axis of the neuroectoderm mirror the organization of gene expression territories along the proximodistal axis of locomotory appendages, including polarized expression of Sp8, Dll, Dac and Hth (Lemons et al., 2010). Similarly, Minelli has suggested that the appendage patterning program could reflect co-option of a more ancient (pre-bilaterian) program for patterning the main body axis and, therefore, bilaterian appendages are simply secondary body axes (Minelli, 2000; Minelli, 2003).
Cephalopod arms and tentacles have no direct structural homologs in non-cephalopod mollusks; however, they likely formed from the ventral embryonic foot, a morphological and embryological hallmark of the molluscan bodyplan (Nödl et al., 2016). Therefore, cephalopod arms and tentacles may be considered evolutionary novelties that are derived from a structure that is conserved across Mollusca. This raises the question of whether other foot-derived outgrowths/appendages (e.g., in sea slugs) evolved by co-option of the same developmental program that cephalopods, arthropods, and vertebrates use to build appendages.
Although the results presented here suggest that an ancient and conserved developmental genetic program facilitated the origin of cephalopod limbs, they also indicate that fine-scale regulatory changes may have played a role in the diversification of cephalopod limb morphologies. For example, evolution of specialized tentacles from serially homologous arms may have resulted from a distal shift in the expression of proximal identity genes, such as Exd and Hth, which could have extended the proximal sucker-free domain and restricted suckers to a distal pad (see Figure 3A–E). Likewise, the results of functional manipulations of Hh signaling in cuttlefish limbs suggests that the diversity in the number of sucker rows in cephalopod limbs (i.e. four rows in squids and cuttlefishes, two in octopus, and one in vampire squid and glass octopus) could be explained by modulation of Hh signaling, in the same way that gradual changes to Shh regulation has led to variation in digit number in tetrapod vertebrates (Scherz et al., 2007; Shapiro et al., 2003; Zhu et al., 2008).
Finally, we note that while the data presented here point to the existence of a deeply conserved genetic program for appendage development across Bilateria, this does not imply that the limbs of cephalopods, arthropods, and vertebrates are homologous structures, or that limbs were present in the common ancestor. Rather, these results show that homologous developmental mechanisms underlie the multiple parallel origins of limbs in bilaterians.
Materials and methods
No statistical methods were used to predetermine sample size. Embryos were randomized in each experiment. The investigators were not blinded to allocation during experiments and outcome assessment.
Embryo collection and preparation
Sepia officinalis and Sepia bandensis eggs were purchased from commercial suppliers, incubated until they reached the required stages (Lemaire, 1970), and prepared for in situ hybridization (ISH) and immunohistochemistry as described (Tarazona et al., 2016).
Optical projection tomography (OPT)
Three-dimensional reconstructions of gene expression in cuttlefish embryos were performed as previously described (Tarazona et al., 2016).
Scanning electron microscopy
Cuttlefish embryos were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) overnight at 4°C and were washed with PBS the next day. Embryos were fixed in 1% osmium tetroxide solution in PBS for 30 min and then washed three times in PBS, dehydrated through a graded ethanol series, critical point dried, and sputter coated with gold. Embryonic samples were scanned using a Hitachi SU5000 and Hitachi TM3000.
Gene cloning and molecular phylogenetic analysis
RNA extraction from Sepia officinalis and Sepia bandensis embryos at stages 15–26 was performed using TRIzol reagent (Ambion) following the manufacturer’s instructions. cDNA synthesis was performed by an AMV reverse transcriptase (New England Biolabs) following the manufacturer’s instructions. PCR amplification was carried out on Sepia cDNA pools, amplicons were cloned into TA vectors and sequenced. We then performed multiple sequence alignments (MSA) with ClustalW (PMID: 7984417) using the predicted amino acid sequence of our cuttlefish cDNA fragments, and putative metazoan orthologous genes downloaded from NCBI RefSeq protein databases (Figure 2—source data 1). We performed nine MSA for Wnt, Tcf, Sfrp, Notum, Patch, Hh, Bmp, Sp and Homeodomain families. Each of the nine MSA was analyzed by ProtTest (PMID: 15647292), in order to determine the best combination of amino acid substitution model and other free parameters (amino acid site frequency, site heterogeneity and invariant sites), using Akaike information criterion (Figure 2—source data 1). We applied the best model in RaXML (PMID: 18853362) for each MSA and performed maximum likelihood phylogenetic inference, estimating branch support by bootstrap, and then majority consensus of the trees from all bootstrap partitions was performed to compute the final tree topology. All sequences have been deposited in Genbank under accession numbers MK756067-MK756082 (complete list of entries is provided in Figure 2—source data 1).
In situ hybridization (ISH) and immunohistochemistry
Whole-mount ISH was performed using digoxigenin- and fluorescein-labeled antisense (or sense control) RNA probes according to protocols described previously (Tarazona et al., 2016). Due to limited availability of embryonic material at relevant early developmental stages, only a limited number of S. bandensis embryos were used for ISH. Thus, the majority of ISH were performed in S. officinalis embryos using S. officinalis antisense RNA probes, however, some ISH were performed in S. officinalis embryos using S. bandensis antisense RNA probes. We validated the specificity of S. bandensis probes in S. officinalis embryos by comparing the gene expression domains marked by these probes in embryonic material from both species at stages 20 and 21. This comparison shows that gene expression territories identified by these probes at these stages were indistinguishable between the two species (Figure 3—figure supplement 2), consistent with their high level of sequence similarity (Figure 3—source data 1). Excluding the S. bandensis ISH mentioned above, all the experiments described in this work were carried out with S. officinalis embryos. Proliferating cells were detected by immunolocalization of Histone H3 Serine 10 phosphorylation using an antibody against H3S10p/PHH3 (06–570, EMD Millipore) and brachial nerve tissue was detected using an antibody against acetylated alpha tubulin (ab24610, Abcam).
Cuttlefish ex-ovo embryo culture and embryo manipulations
A protocol for ex-ovo cuttlefish embryo culture was established for this study, as a modified version of previous descriptions of ex-ovo embryo culture in squid (Arnold, 1990). Briefly, to minimize the problem of bacterial and fungal contamination we started the protocol by taking 10 cuttlefish eggs at the appropriate stage, placing them in a 50 ml tube, and washing them with 0.22 μm filtered artificial sea water (FASW) five times. Eggs were then cleaned with a freshly prepared 5% bleach solution (0.25% sodium hypochlorite in FASW) for 5 s and immediately washed with FASW five times. The bleaching and washing steps were repeated two to three times. Five additional washes with FASW were carried out before incubating the eggs in 2X antibiotic/antimycotic solution (A5955, Sigma) in FASW for 2 hr at ambient temperature.
Each cuttlefish egg was then transferred to a 50 mm diameter petri dish that was coated with a ~ 5 mm layer of 0.5% low melting point agarose (16520050, ThermoFisher), and filled with culture medium (components described below). The agarose layer had a hemispherical depression in the center of the dish made with a sterile 10 mm acrylic necklace bead before gel solidification. The 10 mm hemispherical depression is essential to maintain the normal shape of the yolk mass once the embryos are outside their egg case. Embryos were then extracted from their egg cases (S. officinalis are housed individually, one embryo per egg case) very slowly and with extreme care to avoid rupturing the yolk mass at the vegetal pole of the egg and were carefully placed in the hemispherical depression in the agarose. To extract the embryo, a single 5 mm diameter hole was created in the egg case, which generates a burst of the vitelline liquid and part of the embryo out from the egg case. With the hole kept open, the spontaneous shrinkage of the egg case aided in the expelling of the large cuttlefish embryo. Of every ten eggs prepared this way, between two and five embryos were damaged and had to be discarded. Embryos were cultured at 17°C.
Protein carrier beads and tissue grafting
For protein carrier bead implantation, 150 μm diameter Affi-Gel Blue Gel beads (153–7301, Biorad) were selected and transferred to 1 mg/ml recombinant human Noggin protein (6057 NG, R and D Systems) in PBS and incubated for 30 min to 1 hr at ambient temperature before being implanted in embryos. Control beads were incubated in PBS only.
Grafts of Hh-expressing tissue were performed using stage 24 donor embryos and carefully dissecting the funnel side of the mantle-funnel locking system, which carries the Hh-expressing thickened funnel epithelium (Tarazona et al., 2016). The dissected tissue was transferred to 10 mg/ml Dispase II (D4693, Sigma) in cuttlefish culture medium and incubated for 40 min or until the thickened epithelium was easily detaching from the underlying mesenchyme with the aid of forceps. Tissue was then transferred to cuttlefish culture medium without Dispase II, where they were washed and then grafted into limb buds of stage 17 host embryos. Control grafts were performed using the non-Hh expressing epithelium of the funnel.
After bead implantation or tissue grafts, embryos were incubated at 17°C until control embryos reached stage 26, at which point all embryos were collected and prepared for SEM or ISH.
Cuttlefish culture medium
We used a modified version of a cell culture medium for squid neuron, glia and muscle cells that was previously described (Rice et al., 1990). Cuttlefish culture medium had no glucose, was buffered with 20 mM HEPES and adjusted the pH to 7.6. The medium contained: 430 mM NaCl, 10 mM KCl, 10 mM CaCl2, 50 mM MgCl2, 1X MEM Non-Essential Amino Acids Solution (11140–076, Life Technologies), 1X MEM Amino Acids Solution (11130–051, Life Technologies), 1X MEM Vitamin Solution (11120–052, Life Technologies), 2 mM L-Glutamine (25030–081, Life Technologies). The medium was supplemented with 20% heat inactivated fetal bovine serum (16000044, ThermoFisher) and 1X antibiotic/antimycotic solution (A5955, Sigma).
Treatments with small-molecule inhibitors
Cyclopamine treatments were performed as described previously (Tarazona et al., 2016) with the following modifications; stage 16 embryos were treated with 10 μM cyclopamine (C988400, Toronto Research Chemicals) for 2 days, then washed thoroughly ten times with FASW. Embryos were then washed five more times every hour and one time every day before collecting the embryos for SEM. Control embryos were treated with 0.1% DMSO and then washed as described above.
References
- BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. K Ahn, Y Mishina, MC Hanks, RR Behringer, EB Crenshaw. (2001) Development 128:4449–4461. PubMed, Google Scholar.
- Insect appendages and comparative ontogenetics. DR Angelini, TC Kaufman. (2005) Developmental Biology 286:57–77. PubMed, Google Scholar.
- Normal embryonic stages of the squid, loligo pealii (LESUEUR). JM Arnold. (1965) The Biological Bulletin 128:24–32. Google Scholar.
- Embryonic Development of the Squid. JM Arnold. (1990) In: W. J Adelman, J. M Arnold, D. L Gilbert, editors. Squid as Experimental Animals. Springer. pp. 77–90. Google Scholar.
- Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. K Basler, G Struhl. (1994) Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein PubMed, Google Scholar.
- The Short antennae gene of Tribolium is required for limb development and encodes the orthologue of the Drosophila Distal-less protein. A Beermann, DG Jay, RW Beeman, M Hülskamp, D Tautz, G Jürgens. (2001) The Short antennae gene of Tribolium is required for limb development and encodes the orthologue of the Drosophila Distal-less protein PubMed, Google Scholar.
- Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. R Bolognesi, A Beermann, L Farzana, N Wittkopp, R Lutz, G Balavoine, SJ Brown, R Schröder. (2008) Development Genes and Evolution 218:193–202. PubMed, Google Scholar.
- Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila Leg. WJ Brook, SM Cohen. (1996) Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila Leg PubMed, Google Scholar.
- Embryonic expression of the single Tribolium engrailed homolog. SJ Brown, NH Patel, RE Denell. (1994) Developmental Genetics 15:7–18. PubMed, Google Scholar.
- Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. TD Capellini, G Di Giacomo, V Salsi, A Brendolan, E Ferretti, D Srivastava, V Zappavigna, L Selleri. (2006) Development 133:2263–2273. PubMed, Google Scholar.
- Bmp2, Bmp4 and Bmp7 are co-required in the mouse AER for normal digit patterning but not limb outgrowth. KS Choi, C Lee, DM Maatouk, BD Harfe. (2012) PLOS ONE 7:e37826. PubMed, Google Scholar.
- Development of the external genitalia: conserved and divergent mechanisms of appendage patterning. MJ Cohn. (2011) Developmental Dynamics 240:1108–1115. PubMed, Google Scholar.
- Novel regulatory interactions revealed by studies of murine limb pattern in Wnt-7a and En-1 mutants. JA Cygan, RL Johnson, AP McMahon. (1997) Development 124:5021–5032. PubMed, Google Scholar.
- Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. WG Damen. (2002) Development 129:1239–1250. PubMed, Google Scholar.
- Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. FJ Diaz-Benjumea, B Cohen, SM Cohen. (1994) Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs PubMed, Google Scholar.
- Embryonic development of the cricket Gryllus bimaculatus. S Donoughe, CG Extavour. (2016) Developmental Biology 411:140–156. PubMed, Google Scholar.
- Two new targeted alleles for the comprehensive analysis of Meis1 functions in the mouse. M González-Lázaro, A Roselló-Díez, I Delgado, L Carramolino, MA Sanguino, G Giovinazzo, M Torres. (2014) Genesis 52:967–975. PubMed, Google Scholar.
- Evolution of external genitalia: insights from reptilian development. ML Gredler, CE Larkins, F Leal, AK Lewis, AM Herrera, CL Perriton, TJ Sanger, MJ Cohn. (2014) Sexual Development 8:311–326. PubMed, Google Scholar.
- A hedgehog homolog is involved in muscle formation and organization of Sepia officinalis (mollusca) mantle. A Grimaldi, G Tettamanti, F Acquati, E Bossi, ML Guidali, S Banfi, L Monti, R Valvassori, M de Eguileor. (2008) Developmental Dynamics 237:659–671. PubMed, Google Scholar.
- Formation of body appendages during caudal regeneration in Platynereis dumerilii: adaptation of conserved molecular toolsets. J Grimmel, AW Dorresteijn, AC Fröbius. (2016) Formation of body appendages during caudal regeneration in Platynereis dumerilii: adaptation of conserved molecular toolsets PubMed, Google Scholar.
- Gene expression analysis of potential morphogen signalling modifying factors in Panarthropoda. M Hogvall, GE Budd, R Janssen. (2018) EvoDevo 9:20. PubMed, Google Scholar.
- Gene expression suggests decoupled dorsal and ventral segmentation in the millipede Glomeris marginata (Myriapoda: Diplopoda). R Janssen, NM Prpic, WG Damen. (2004) Developmental Biology 268:89–104. PubMed, Google Scholar.
- Complementary and mutually exclusive activities of decapentaplegic and wingless organize axial patterning during Drosophila leg development. J Jiang, G Struhl. (1996) Complementary and mutually exclusive activities of decapentaplegic and wingless organize axial patterning during Drosophila leg development PubMed, Google Scholar.
- Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Y Kawakami, CR Esteban, T Matsui, J Rodríguez-León, S Kato, JC Izpisúa Belmonte. (2004) Development 131:4763–4774. PubMed, Google Scholar.
- Induction of a mirror-image duplication of anterior wing structures by localized hedgehog expression in the anterior compartment of Drosophila melanogaster wing imaginal discs. T Kojima, T Michiue, M Orihara, K Saigo. (1994) Induction of a mirror-image duplication of anterior wing structures by localized hedgehog expression in the anterior compartment of Drosophila melanogaster wing imaginal discs PubMed, Google Scholar.
- Expression and function of the zinc finger transcription factor Sp6-9 in the spider Parasteatoda tepidariorum. T Königsmann, N Turetzek, M Pechmann, NM Prpic. (2017) Development Genes and Evolution 227:389–400. PubMed, Google Scholar.
- Cephalopod origin and evolution: A congruent picture emerging from fossils, development and molecules. B Kröger, J Vinther, D Fuchs. (2011) BioEssays 33:602–613. Google Scholar.
- Proximal-distal axis formation in the Drosophila leg. T Lecuit, SM Cohen. (1997) Proximal-distal axis formation in the Drosophila leg PubMed, Google Scholar.
- Cephalopod Hox genes and the origin of morphological novelties. PN Lee, P Callaerts, HG De Couet, MQ Martindale. (2003) Nature 424:1061–1065. PubMed, Google Scholar.
- Table de developpement embryonnaire de Sepia officinalis. L. (Mollusque Cephalopode). J Lemaire. (1970) Table de developpement embryonnaire de Sepia officinalis. L. (Mollusque Cephalopode) Google Scholar.
- Co-option of an anteroposterior head axis patterning system for proximodistal patterning of appendages in early bilaterian evolution. D Lemons, JH Fritzenwanker, J Gerhart, CJ Lowe, W McGinnis. (2010) Developmental Biology 344:358–362. PubMed, Google Scholar.
- Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. PM Lewis, MP Dunn, JA McMahon, M Logan, JF Martin, B St-Jacques, AP McMahon. (2001) Cell 105:599–612. PubMed, Google Scholar.
- Analysis of the genetic pathway leading to formation of ectopic apical ectodermal ridges in mouse Engrailed-1 mutant limbs. CA Loomis, RA Kimmel, CX Tong, J Michaud, AL Joyner. (1998) Development 125:1137–1148. PubMed, Google Scholar.
- Conserved regulation of proximodistal limb axis development by Meis1/Hth. N Mercader, E Leonardo, N Azpiazu, A Serrano, G Morata, C Martínez, M Torres. (1999) Nature 402:425–429. PubMed, Google Scholar.
- Limbs and tail as evolutionarily diverging duplicates of the main body axis. A Minelli. (2000) Evolution and Development 2:157–165. PubMed, Google Scholar.
- The Development of Animal Form: Ontogeny, Morphology, and Evolution. A Minelli. (2003) Cambridge University Press. Google Scholar.
- Conservation, innovation, and the evolution of horned beetle diversity. AP Moczek, D Rose, W Sewell, BR Kesselring. (2006) Development Genes and Evolution 216:655–665. PubMed, Google Scholar.
- Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. AP Moczek, LM Nagy. (2005) Evolution & Development 7:175–185. PubMed, Google Scholar.
- Animal Evolution: Interrelationships of the Living Phyla. C Nielsen. (2012) Oxford: Oxford press. Google Scholar.
- The cephalopod arm crown: appendage formation and differentiation in the Hawaiian bobtail squid Euprymna scolopes. MT Nödl, A Kerbl, MG Walzl, GB Müller, HG de Couet. (2016) The cephalopod arm crown: appendage formation and differentiation in the Hawaiian bobtail squid Euprymna scolopes PubMed, Google Scholar.
- The origin and evolution of animal appendages. G Panganiban, SM Irvine, C Lowe, H Roehl, LS Corley, B Sherbon, JK Grenier, JF Fallon, J Kimble, M Walker, GA Wray, BJ Swalla, MQ Martindale, SB Carroll. (1997) PNAS 94:5162–5166. PubMed, Google Scholar.
- Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. BA Parr, AP McMahon. (1995) Nature 374:350–353. PubMed, Google Scholar.
- Expression of dachshund in wild-type and Distal-less mutant Tribolium corroborates serial homologies in insect appendages. NM Prpic, B Wigand, WG Damen, M Klingler. (2001) Development Genes and Evolution 211:467–477. PubMed, Google Scholar.
- Gene expression in spider appendages reveals reversal of exd/hth spatial specificity, altered leg gap gene dynamics, and suggests divergent distal morphogen signaling. NM Prpic, R Janssen, B Wigand, M Klingler, WG Damen. (2003) Developmental Biology 264:119–140. PubMed, Google Scholar.
- Homologs of wingless and decapentaplegic display a complex and dynamic expression profile during appendage development in the millipede Glomeris marginata (Myriapoda: Diplopoda). NM Prpic. (2004) Frontiers in Zoology 1:6. PubMed, Google Scholar.
- Parasegmental appendage allocation in annelids and arthropods and the homology of parapodia and arthropodia. NM Prpic. (2008) Frontiers in Zoology 5:17. PubMed, Google Scholar.
- The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. NM Prpic, D Tautz. (2003) Developmental Biology 260:97–112. PubMed, Google Scholar.
- Parallels between the proximal-distal development of vertebrate and arthropod appendages: homology without an ancestor?. JI Pueyo, JP Couso. (2005) Current Opinion in Genetics & Development 15:439–446. PubMed, Google Scholar.
- Tissue culture of squid neurons, glia, and muscle cells. RV Rice, R Mueller, WJ Adelman. (1990) In: D. L Gilbert, W. J Adelman, J. M Arnold, editors. Squids as Experimental Animals. Springer. pp. 195–212. Google Scholar.
- Sonic hedgehog mediates the polarizing activity of the ZPA. RD Riddle, RL Johnson, E Laufer, C Tabin. (1993) Cell 75:1401–1416. PubMed, Google Scholar.
- Invertebrate Zoology: A Functional Evolutionary Approach. EE Ruppert, RS Fox, RD Barnes. (2004) Thomson-Brooks/Cole. Google Scholar.
- Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. V Salsi, MA Vigano, F Cocchiarella, R Mantovani, V Zappavigna. (2008) Developmental Biology 317:497–507. PubMed, Google Scholar.
- The Tribolium decapentaplegic gene is similar in sequence, structure, and expression to the Drosophila dpp gene. J Sanchez-Salazar, MT Pletcher, RL Bennett, SJ Brown, TJ Dandamudi, RE Denell, JS Doctor. (1996) The Tribolium decapentaplegic gene is similar in sequence, structure, and expression to the Drosophila dpp gene PubMed, Google Scholar.
- A clustered set of three Sp-family genes is ancestral in the Metazoa: evidence from sequence analysis, protein domain structure, developmental expression patterns and chromosomal location. ND Schaeper, NM Prpic, EA Wimmer. (2010) BMC Evolutionary Biology 10:88. PubMed, Google Scholar.
- Extended exposure to Sonic hedgehog is required for patterning the posterior digits of the vertebrate limb. PJ Scherz, E McGlinn, S Nissim, CJ Tabin. (2007) Developmental Biology 308:343–354. PubMed, Google Scholar.
- Cooption of an appendage-patterning gene cassette in the head segmentation of arachnids. EVW Setton, PP Sharma. (2018) PNAS 115:E3491–E3500. PubMed, Google Scholar.
- Developmental basis of evolutionary digit loss in the australian lizard Hemiergis. MD Shapiro, J Hanken, N Rosenthal. (2003) Developmental basis of evolutionary digit loss in the australian lizard Hemiergis PubMed, Google Scholar.
- Evolution of the cephalopod head complex by assembly of multiple molluscan body parts: Evidence from Nautilus embryonic development. S Shigeno, T Sasaki, T Moritaki, T Kasugai, M Vecchione, K Agata. (2008) Journal of Morphology 269:1–17. PubMed, Google Scholar.
- Fossils, genes and the evolution of animal limbs. N Shubin, C Tabin, S Carroll. (1997) Nature 388:639–648. PubMed, Google Scholar.
- Deep homology and the origins of evolutionary novelty. N Shubin, C Tabin, S Carroll. (2009) Nature 457:818–823. PubMed, Google Scholar.
- Resolving the evolutionary relationships of molluscs with phylogenomic tools. SA Smith, NG Wilson, FE Goetz, C Feehery, SC Andrade, GW Rouse, G Giribet, CW Dunn. (2011) Nature 480:364–367. PubMed, Google Scholar.
- Out on a limb parallels in vertebrate and invertebrate limb patterning and the origin of appendages. CJ Tabin, SB Carroll, G Panganiban. (1999) American Zoologist 39:650–663. Google Scholar.
- The genetic program for cartilage development has deep homology within Bilateria. OA Tarazona, LA Slota, DH Lopez, G Zhang, MJ Cohn. (2016) Nature 533:86–89. PubMed, Google Scholar.
- BMP-mediated functional cooperation between Dlx5;Dlx6 and Msx1;Msx2 during mammalian limb development. M Vieux-Rochas, K Bouhali, S Mantero, G Garaffo, P Provero, S Astigiano, O Barbieri, MF Caratozzolo, A Tullo, L Guerrini, Y Lallemand, B Robert, G Levi, GR Merlo. (2013) PLOS ONE 8:e51700. PubMed, Google Scholar.
- Expression of Distal-less, dachshund, and optomotor blind in Neanthes arenaceodentata (Annelida, Nereididae) does not support homology of appendage-forming mechanisms across the Bilateria. CJ Winchell, JE Valencia, DK Jacobs. (2010) Development Genes and Evolution 220:275–295. PubMed, Google Scholar.
- Expression of the Lhx genes apterous and lim1 in an errant polychaete: implications for bilaterian appendage evolution, neural development, and muscle diversification. CJ Winchell, DK Jacobs. (2013) EvoDevo 4:4, 4. Google Scholar.
- Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. F Witte, J Dokas, F Neuendorf, S Mundlos, S Stricker. (2009) Gene Expression Patterns 9:215–223. PubMed, Google Scholar.
- Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. J Zhu, E Nakamura, MT Nguyen, X Bao, H Akiyama, S Mackem. (2008) Developmental Cell 14:624–632. PubMed, Google Scholar.
Decision letter
-
Nikola-Michael Prpic-SchäperReviewing Editor; University of Giessen, Germany
-
Diethard TautzSenior Editor; Max-Planck Institute for Evolutionary Biology, Germany
-
Gunter WagnerReviewer; Yale University, United States
In the interests of transparency, eLife includes the editorial decision letter and accompanying author responses. A lightly edited version of the letter sent to the authors after peer review is shown, indicating the most substantive concerns; minor comments are not usually included.
Thank you for submitting your work entitled "Evolution of limb development in cephalopod mollusks" for consideration at eLife. Your article has been favorably evaluated by Diethard Tautz (Senior Editor), a Reviewing Editor, and two reviewers.
The reviewers have discussed the reviews with one another and the Reviewing Editor has drafted this decision to help you prepare a revised submission.
1) Please consider extending the Discussion along the following guidelines:
a) Consider discussing a possible ancestral function of the conserved patterning module identified in your work, if the original function was not limb patterning (suggested by reviewer #1). Parts of the gene patterning functions are actually found in phylogenetically older animal groups: could these mirror aspects of the original function of the patterning module? (suggested by reviewer #2).
b) Consider including a schematical summary of the gene expression patterns in the cephalopod compared with vertebrates and arthropods, to make it easier for readers to appreciate the similarities between these animal groups (suggested by reviewer #2).
2) In most of the phylogenetic trees shown in the supplementary data there are the sequences for S. bandensis and not S. officinalis. Please clarify why there are sequences from two different species, when the cloning and further analysis was only carried out in S. officinalis. (suggested by reviewer #2).
3) Please also revise the following points (combined from the suggestions by both reviewers):
a) You mention in your manuscript that there are no other mollusks that have appendages. Could the captacula of Scaphopods be homologous to cephalopod tentacles? If so, this could be worth mentioning.
b) Discussion: expression patterns of hth, exd, Dll, and dpp can be found for Platynereis during caudal regeneration in Grimmel et al., 2016 (doi: 10.1186/s13227-016-0046-6).
c) Figure 1G: the legend for the DNA counterstaining should be called Hoechst.
d) Figure 2: each panel of the composite should have its own label (same for Figure 3F-I). For better overview, all limb buds should be shown in the same orientation. In some in situ pictures it is quite hard to see the general shape of the limb buds. Here, a dotted outline would make the pictures much clearer (similar to Figure 4H) (same for Figure 3 F-I).
Please find below the full comments by both reviewers, for your information:
Reviewer #1:
I congratulate the authors to an outstanding contribution to evolutionary developmental biology. The authors demonstrate that cephalopod appendages, arms and tentacles, are patterned by a gene regulatory network that is homologous to those active in vertebrate fin/limb buds and arthropod legs. The authors make their case through examination of the RNA expression patterns of key members of the appendage gene regulatory network as well as experimental manipulation of the dorso-ventral and the anterior-posterior axis, yielding results consistent with what is known from vertebrate and arthropod appendages. The authors do carefully consider and test alternative interpretations of their findings, and I find the corpus of evidence convincing. These findings support the notion that there is a patterning module that existed in the most recent common ancestor of bilaterians and which has been inherited to "limbed" members of all major bilaterian clades.
As all important discoveries, this too raises new questions that the authors may want to consider more extensively than done in the submitted manuscripts. If, in fact, the limb patterning module is as old as or rather older than the most recent common ancestor of bilaterians this module must have played a developmental role in that ancestor. The authors say that their findings do not imply that this ancestor had "limbs" but then what was it doing? Also, there has to be a function for this module that explains its conservation from the bilaterian ancestor to the modern groups which have, independently, evolved limbs. Is there a non-limb/appendage function known of this patterning module, or do we have to assume a massive loss of appendages in the evolution of the non-limbed crown bilaterians?
Reviewer #2:
The authors describe the molecular mechanisms underlying the development of appendages in a cephalopod mollusk. The article focuses on the expression and function of genes involved in limb development known from arthropods and vertebrates and thus presents the first comprehensive analysis of appendage formation in Spiralia.
The authors state convincingly, that the spatio-temporal expression patterns of the genes analysed show conserved aspects to the known determination networks, as well as the function of Bmp and Hh signalling. They also propose a molecular reason for the morphological difference between arms and tentacles.
From their findings the authors conclude, that not the bilaterian limbs are homologous, but rather their underlying developmental mechanisms.
The in situ data are of acceptable quality, clearly presented and the manuscript is very well written.
The authors state, that expression patterns along the proximodistal axis are "similarly polarized" when compared to arthropods and vertebrates. However, especially in the case of exd and hth the expression patterns vary among arthropods. Similarly, dac usually marks the median part of an outgrowing limb in arthropods and is not restricted to the distal part. For the dorsoventral axis specification, the article says that dpp is expressed dorsally in arthropods. While this is true for Drosophila, expression patterns of dpp in other arthropods are strikingly different and not thought to be ancestrally involved in axis formation (see Angelini and Kaufman, 2005, for an overview of expression patterns in arthropods). The existence of these all differences between arthropods, vertebrates and mollusks in expression patterns of the genes analysed should be mentioned and discussed. To this end it would be advantageous to depict a schematic representation of the various expression patterns in the three animal groups to aid readers during the discussion.
Additionally, since the authors come to the conclusion that the developmental mechanisms are homologous between the different animal groups. The Discussion should mention the presence of parts of these mechanisms in non-bilaterian groups. For instance, differential expression of wnt in the a-p axis of sponge embryos, or the establishment of the oral-aboral axis in cnidarians.
Finally, in most of the phylogenetic trees shown in the supplementary data there are the sequences for S. bandensis and not S. officinalis. Please clarify why there are sequences from two different species, when the cloning and further analysis was only carried out in S. officinalis.
https://doi.org/10.7554/eLife.43828.031Author response
The reviewers have discussed the reviews with one another and the Reviewing Editor has drafted this decision to help you prepare a revised submission.
1) Please consider extending the Discussion along the following guidelines:
a) Consider discussing a possible ancestral function of the conserved patterning module identified in your work, if the original function was not limb patterning (suggested by reviewer #1). Parts of the gene patterning functions are actually found in phylogenetically older animal groups: could these mirror aspects of the original function of the patterning module? (suggested by reviewer #2).
We have added a discussion of the possible ancestral role(s) of the genetic circuit that controls appendage development (Discussion paragraphs three, four and five). We propose that this developmental program might have its roots in the development of non-locomotory appendages (outgrowths from the primary body axis) in other metazoans, including those that were limbless. We also note that these signaling pathways are known to function elsewhere in the bodies of vertebrates, arthropods, and cephalopods. We refer to two previously proposed hypothesis that are consistent with the idea that this genetic circuit originally patterned the anteroposterior axis of the trunk (or the cephalic neuroectoderm) in bilaterians and was repeatedly co-opted to perform the same function (axial patterning) in secondary outgrowths, including locomotory appendages (Discussion paragraph three).
The Discussion also addresses the implications of our finding that these genes are expressed in the developing funnel/siphon of cephalopods (paragraph four). In a new supplemental figure (Figure 5—figure supplement 1), we show that Wnt5 and Exd, which are involved in proximal-distal limb patterning, also are expressed in the funnel/siphon, and the patterns that mirror their expression domains in the limbs. We discuss the possibility that the same genetic circuit could control funnel/siphon and limb development, and that its role in funnel development could potentially predate the origin of cephalopod limbs. Therefore, in the broader context of animal development, independent evolution of appendages could be explained by the parallel redeployment of an ancient developmental genetic program that predates the structures themselves and very likely controlled the development of non-limb outgrowths, such as vertebrate genitalia, beetle horns, or mollusk siphon. Accordingly, application of the term “limbs” to cephalopod arms and tentacles does not make them any more closely related to vertebrate limbs than to vertebrate genitalia or any other animal appendage.
b) Consider including a schematical summary of the gene expression patterns in the cephalopod compared with vertebrates and arthropods, to make it easier for readers to appreciate the similarities between these animal groups (suggested by reviewer #2).
Thank you for this suggestion. We have created a new schematic figure (Figure 5) that compares gene expression in vertebrate, arthropod, and cephalopod limbs. This schematic figure shows the conserved and the variable patterns of gene expression for most of the genes analyzed in our paper compared to published expression patterns in arthropods and vertebrates. The references used for the construction of this figure are cited in the figure legend.
2) In most of the phylogenetic trees shown in the supplementary data there are the sequences for S. bandensis and not S. officinalis. Please clarify why there are sequences from two different species, when the cloning and further analysis was only carried out in S. officinalis. (suggested by reviewer #2).
We have clarified in the main text that while most of the gene cloning was performed in Sepia officinalis, some clones were isolated from Sepia bandensis, a closely related species. We show that the S. officinalis and S. bandensis clones have high sequence similarity (Figure 3—figure supplement 1), and comparison of mRNA localization in S. officinalis and S. bandensis embryos by in situ hybridization showed that the expression patterns in the developing limbs are indistinguishable in the two species. These new data are shown in a new supplementary figure and legend (Figure 3—figure supplement 2).
3) Please also revise the following points (combined from the suggestions by both reviewers):
a) You mention in your manuscript that there are no other mollusks that have appendages. Could the captacula of Scaphopods be homologous to cephalopod tentacles? If so, this could be worth mentioning.
We are unaware of evidence supporting structural homology of scaphopod captacula and cephalopod tentacles. It appears that these appendages have different embryonic origins, which would suggest that they are not homologous structures (sensu, derived from a common ancestral appendage). To the larger point, we have added a discussion of polychaete appendages and we propose that the gene regulatory network that we identified in cephalopod arms and tentacles plays a general and conserved role in bilaterian appendage development, irrespective of the type of appendage (Discussion, second paragraph).
b) Discussion: expression patterns of hth, exd, Dll, and dpp can be found for Platynereis during caudal regeneration in Grimmel et al., 2016 (doi: 10.1186/s13227-016-0046-6).
We have added this reference and discussed its implications for parapodia and for the broader question of appendage evolution in Bilateria.
c) Figure 1G: the legend for the DNA counterstaining should be called Hoechst.
Thank you. This has been added to the legend.
d) Figure 2: each panel of the composite should have its own label (same for Figure 3F-I). For better overview, all limb buds should be shown in the same orientation. In some in situ pictures it is quite hard to see the general shape of the limb buds. Here, a dotted outline would make the pictures much clearer (similar to Figure 4H) (same for Figure 3 F-I).
We have labeled the stage on every panel. However, we have retained the orientation of the limbs in order to enable readers to easily see the differences that we describe for cephalopod limbs. This is particularly important for the expression of Hh and Patched, because orienting them in the same way as the other limb buds could inadvertently lead readers to misinterpret the pattern as dorsoventrally polarized instead of anteriorly polarized. We have included axis labels in every panel as an additional tool to help readers with the interpretation of the spatial domains.
https://doi.org/10.7554/eLife.43828.032Article and author information
Author details
-
Oscar A Tarazona
- Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, United States
- Department of Biology, UF Genetics Institute, University of Florida, Gainesville, United States
Contribution
Conceptualization, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing—original draftCompeting interests
No competing interests declared -
Davys H Lopez
Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, United States Contribution
Validation, InvestigationCompeting interests
No competing interests declared -
Leslie A Slota
Department of Biology, UF Genetics Institute, University of Florida, Gainesville, United States Contribution
Validation, InvestigationCompeting interests
No competing interests declaredORCID iD: 0000-0001-6911-811X -
Martin J Cohn
- Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, United States
- Department of Biology, UF Genetics Institute, University of Florida, Gainesville, United States
Contribution
Conceptualization, Resources, Supervision, Funding acquisition, Validation, Project administration, Writing—review and editingFor correspondence
mjcohn@ufl.eduCompeting interests
No competing interests declaredORCID iD: 0000-0002-5211-200X
Funding
Howard Hughes Medical Institute
- Oscar A Tarazona
- Davys H Lopez
- Leslie A Slota
- Martin J Cohn
Howard Hughes Medical Institute (International Student Research Fellowship)
- Oscar A Tarazona
Society for Developmental Biology ('Choose Development!' fellowship)
- Davys H Lopez
National Science Foundation (EDEN Undergraduate Internship)
- Leslie A Slota
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Emily Merton for technical support, Karen L Kelley and Kimberly L Backer-Kelley (UF ICBR) for assistance with electron microscopy, and members of our laboratory for helpful comments and discussions. OAT was supported by a Howard Hughes Medical Institute International Student Research Fellowship, DHL by a Society for Developmental Biology ‘Choose Development!’ fellowship, and LAS by an EDEN Undergraduate Internship.
Senior Editor
- Diethard Tautz, Max-Planck Institute for Evolutionary Biology, Germany
Reviewing Editor
- Nikola-Michael Prpic-Schäper, University of Giessen, Germany
Reviewer
- Gunter Wagner, Yale University, United States
Publication history
- Received: November 22, 2018
- Accepted: May 8, 2019
- Version of Record published: June 18, 2019 (version 1)
Copyright
© 2019, Tarazona et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Download links
Downloads (link to download the article as PDF)
Download citations (links to download the citations from this article in formats compatible with various reference manager tools)
Open citations (links to open the citations from this article in various online reference manager services)
Related to
Limb Development: A lesson in homology
Further reading
-
- Developmental Biology
- Evolutionary Biology
Nikola-Michael PrpicThe same genes and signaling pathways control the formation of limbs in vertebrates, arthropods and cuttlefish.
-
- Developmental Biology
Noriko Ichino, Stephen C Ekker -
- Cell Biology
- Developmental Biology
Péter Lőrincz et al.