Discovery of scientific software

TL;DR: An open API for science helps researchers discover great software. Install the Scholar Ninja extension and you’ll get recommendations (based on software citations) on-the-fly while browsing GitHub.
A while back I wrote about an open distributed search engine for science, Scholar Ninja, and about how great it will be to have an open API which you can query and get to all of science, no matter if you’re human or machine. Having the world’s knowledge openly accessible like that will result in a paradigm shift. I dare you to say it ain’t so! (Also, check out ContentMine!)
While that project is still in early stages of development (most recently, we even had to turn off our core feature, WebRTC), lots of people have asked what the use cases for such a search engine/API would be, anyway. A great many don’t appreciate how important this will be, and while that might seem silly at first glance, the vast majority of researchers is quite satisfied with the closed, machine-unfriendly Google Scholar and other closed behemoths. How can we show them the light?
After a lot of conversations at the #OKFest14 and more recently at the Mozilla Science Lab Code Sprint, one use case that surfaced from virtually every discussion, was helping scientists (or anyone) discover great software in their field (or any field, or anywhere!). As a tsunami of code is reaching the shores of Science, discovering scientific software will only get more important:
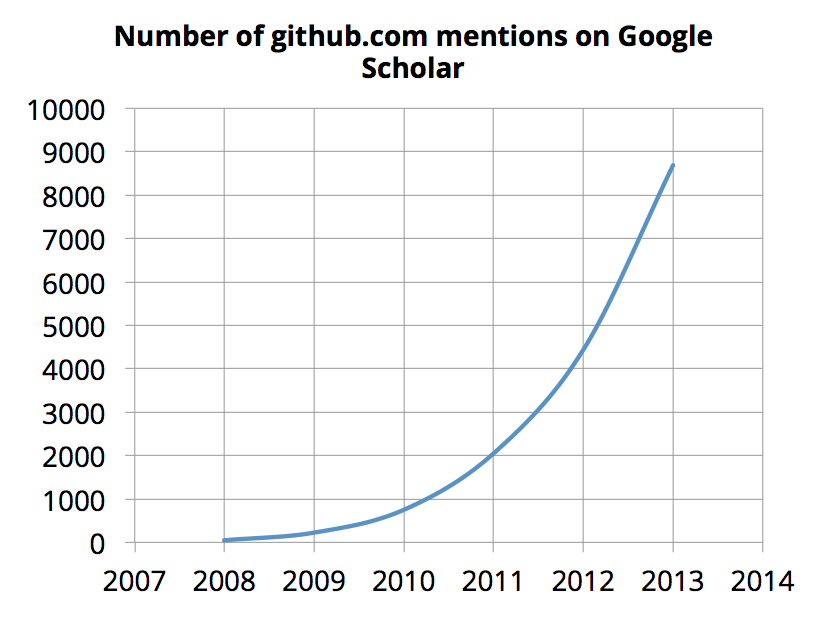
Luckily, it’s quite possible to use Scholar Ninja for scientific software discovery. In fact, it’s more than possible — it’s already done.
Before we get ahead of ourselves, let me provide just a bit of necessary backstory: Scholar Ninja indexes (or rather, did and will again index, see issues) every paper you read online and adds paper’s metadata, keywords and URLs to a globally distributed search index, which is based on browsers, WebRTC and magic. Everyone who has the extension installed is a node in a Chord DHT network and is both an indexer and a server of content. Scholar Ninja’s mission in life is to become a complete and completely open search engine for science.
To get right back to the main story here — if we can get URLs from each paper, that means we can get a large portion of software citations, which usually look something like this:
... reads longer than 300 bases were separated by barcode and trimmed using sickle (https://github.com/najoshi/sickle); 72.3% of reads were retained ...
That is, URL inlined directly into the text. Or referenced classically in the References section:
24. Nikhil J. Sickle - a windowed adaptive trimming tool for FASTQ files using quality. https://github.com/najoshi/sickle.
In both cases the URL is right there, and that appears to be case for most software citations. The URL itself usually points to places like GitHub, BitBucket, SourceForge, Google Code, R Project, etc.
Alright, enough mumbling, imagine that you’re new to the field of bioinformatics and your first task on the job is to process a vast number of FASTQ files representing partial genetic sequences and assemble them into a coherent sequence. “No problem”, you say, “first I need to analyse the sequences and trim them when their quality gets too low.” You start to do this by hand and a week goes by, then two, then three weeks, woosh. Luckily a wise co-worker walks by your tiny windowless office, sees what you’re doing, chuckles kindly, then proceeds to tell you about this great piece of software called sickle and gives you a link to the GitHub repository. Now, naturally, you find sickle fantastic and wonder how you ever could have processed your FASTQ files without its windowed adaptive trimming functionality. “Sickle is the best!” You’d like to know if there are more tools like it out there that could save you hours and days and weeks of work in the future. Unfortunately your co-worker just left the department to go sailing around the world for a year, so you have no one to ask. What can you do?
Here is where the power of an open API for science can really start to shine: How about you take a weekend and create a scientific software recommender system based on software citations that you get from the API?
Let’s do it!
Step 0. Create open API for science
This is what Scholar Ninja hopes to provide in the future. It’s a sizeable project, fraught with many perils, not least of which are the many licensing restrictions publishers place on scientific content. But I digress. We’ve had a working version of the network running with around 70 nodes at peak, but due to bugs in Chrome and our own implementation, it turned out to not be ready for prime time just yet. However, once complete, if you’ll run a Scholar Ninja super-node in Node.js, you’ll be able to join the network and query it just like you would query a good old regular HTTP API.
Step 1. Use open API for science
OK, step 0 just gave you a great API to work with. For our purpose, you could for example ask it to give you all citations of sickle:
GET /citations/software?url=github.com/najoshi/sickle
And you'll get a nice JSON back, with the papers that cite sickle. OK, so now query the API to give you other software which these papers cite:
GET /software?cited_in=["10.1155/2014/404578", "10.1371/journal.pone.0101021", ...]
And again get a nice JSON back:
[{
"id": "github.com/jstjohn/SeqPrep",
"source": "doi:10.1155/2014/404578",
},
{
"id": "github.com/vsbuffalo/scythe",
"source": "doi:10.1186/gb-2013-14-6-r66",
},
...]
Well that was easy. We have our data, all we need to do now is package it up and present it to the user.
Step 2. Win.
For the purposes of this demonstration, I’ve indexed 848,418 open access papers from Europe PMC and analysed 18,765,516 citations, plus 10,837 software citations found within (source code here). When Scholar Ninja’s network will be fully operational, the indexing will be real-time and organic, as users navigate the web and read scientific papers — but have patience, we’re not there yet. However, to help pass the time, if you install the extension right now, every GitHub page where scientific software is detected and recommendations are available, will be enhanced by a neat unobtrusive panel full of great scientific software:
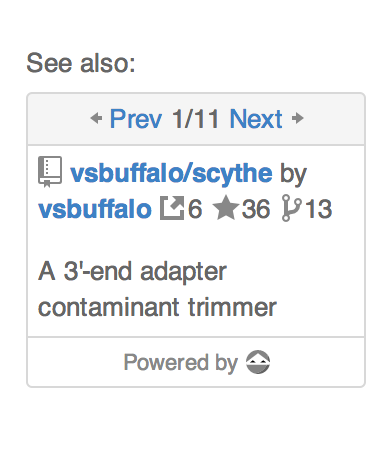
There are eleven recommendations for https://github.com/najoshi/sickle. Eleven pieces of software were cited in papers where sickle was cited and in papers which the paper where sickle was cited cited. Wooh. The external link icon above signifies citations, for example, we found 6 citations for scythe. The rest should be self explanatory. This view is then simply embedded into the standard GitHub interface:
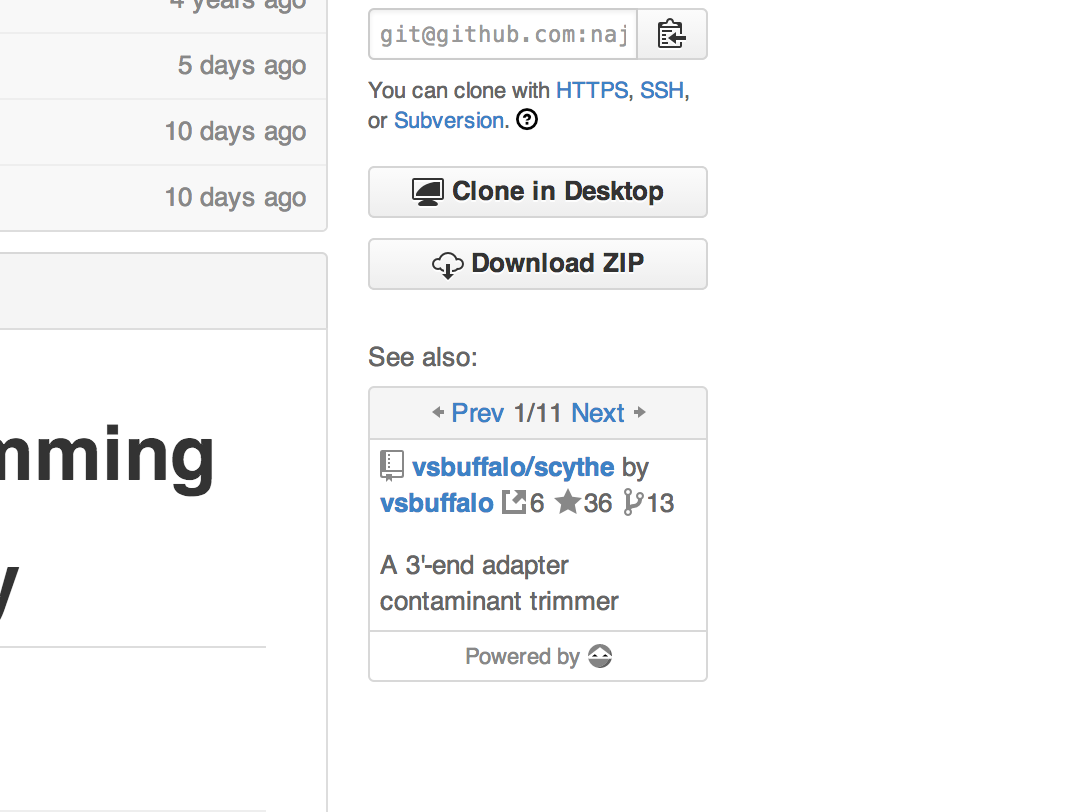
Awesome? I sure think so. Be sure to install the extension and try it out! And this is just a very very tiny glimpse of what the future holds if we build an open API for science. Come on, join us!

